Bromenshenk
TABLE OF CONTENTS
What Do Bromenshenk and the Army Claim to Have Found?
Bromenshenk
Randy Oliver
ScientificBeekeeping.com
First published in American Bee Journal December 2010
A man walking along the street one night sees another searching for his lost keys under a street lamp and politely stops to help. After a long search, the passerby says, “Are you sure this is where you lost the keys?”
“Oh no,” answers the other, “I don’t know where I lost them.”
“Then why are you looking only under the street lamp?
“Because the light’s better here.”
A recent paper, “Iridovirus and Microsporidian Linked to Honey Bee Colony Decline,” by Dr. Jerry Bromenshenk and a team of collaborators (including the U.S. Army’s Edgewood Chemical Biological Center) made national news in early October. Has the Bromenshenk team finally found another clue to the mystery of CCD by looking somewhere that others previously hadn’t? I will interrupt my “Sick Bees” series in order to discuss the implications of this paper.
THE PAPER
I’m writing this article shortly after the release of the paper, and things are crazy! The New York Times ran the unfortunate, but attention-grabbing headline “Scientists and Soldiers Solve a Bee Mystery,” which set off a flurry of excitement in a public worried about honey bees. There is gratuitous sniping from other researchers in the press, and Fortune magazine carried an inaccurate and libelous piece accusing Bromenshenk of being in Bayer’s pocket because he didn’t blame CCD on the neonicotinoid pesticides (as though the Army lab could care one whit about Bayer)! I’m incredulous that folk are getting so worked up! Science shouldn’t be about personalities and politics; it should be about assessing the data and conclusions on their merits.
I want to make clear at this point that I have no dog is this fight, and am as curious as anyone to see if Bromenshenk’s results will be confirmed by other investigators. I know most of the CCD researchers, and find every one of them to be top-notch scientists who really know their bees. So I am going to avoid any political commentary on the progression of CCD research.
I do recommend, that in order to appreciate the scope of the scientific effort to address CCD, that you download the CCD Action Plan (Hackett 2007), which I find to be thorough, meticulous, and impressive (although surprisingly lacking in any suggestion to use the Army lab’s tools, despite its commander being on the Steering Committee). I can assure you that all researchers have diligently spent long hours doing the painstaking and tedious field and lab work necessary to figure out what is going on in the sick hives. It has been as frustrating to them as it has been to struggling beekeepers begging for answers.
WHAT DO BROMENSHENK AND THE ARMY CLAIM TO HAVE FOUND?
Let’s briefly go over the paper section by section—I assume that you’ve downloaded your copy (see References). The paper has its critics, a number of whom I have spoken with; I will attempt to address their criticisms in my analysis. Let’s start with the authors’ summary in the Abstract:
“We used Mass spectrometry-based proteomics (MSP) to identify and quantify thousands of proteins from healthy and collapsing bee colonies. MSP revealed two unreported RNA viruses in North American honey bees, Varroa destructor virus-1 and Kakugo virus, and identified an invertebrate iridescent virus (IIV) (Iridoviridae) associated with CCD colonies.”
Both VDV-1 and Kakugo virus are very closely related to Deformed Wing Virus (DWV), which is ubiquitous in U.S. bees that are infested with varroa. Due to the close relationship to DWV, the identity of these two viruses will need to be confirmed by other methods. If indeed they are present, it will be a wakeup call as to just how porous our borders are to the influx of new bee pathogens!
The key (and most surprising) finding was that the team discovered the apparent widespread presence of a heretofore unreported iridescent virus in sick colonies. This finding has been strongly questioned by some—I will review the evidence shortly. Bromenshenk’s next conclusion is much less controversial:
“In addition, bees in failing colonies contained not only IIV, but also Nosema….We conclude that the IIV/Nosema association may be critical in honey bee mortality linked to CCD.”
The nosema has been subsequently identified as Nosema ceranae (Bromenshenk, pers comm). This may be an “aha!” finding, as bee health problems in a number of countries appear to have increased at about the same time as N. ceranae was introduced, yet it has been hard to pin collapses on N. ceranae alone.
THE TEAM
Bromenshenk teamed up with statistician Dr. Colin Henderson, and then enlisted the help of virologists and chemists at the Army’s Edgewood lab – this is the outfit whose job it is to identify the cause of any new diseases suffered by soldiers (and more recently, civilians and agriculture as well). The Army team was headed by Dr. Evan Skowronski (who, due to being on the CCD steering committee properly recused himself from the actual research). The sample analyses was headed by the Army’s Dr. Charles Wick, who invented the IVDS machine (his brother Dave offers IVDS sampling to beekeepers), who coordinated several additional software designers. When the team found N. ceranae and IIV, they recruited fungal pathologist Dr. Robert Cramer, and iridovirus experts Dr. Shan Bilimoria (who recently patented an IIV protein for use as an insecticide), and Dr. Trevor Williams (who wrote the books on iridoviruses).
THE TOOLS
In order to grasp the significance of the Bromenshenk paper, one must understand the various methods available for confirming the presence of a pathogen in, say, a bee (Figure 1):
- Microscopy (optical or electron) – with which you visually look for the actual organism, as when counting nosema spores.
- Integrated Virus Detection System (IVDS) – this machine separates out viruses by particle size (Dave Wick has recently matched several peaks to specific bee viruses)
- Immunological – in which you use an antibody designed to bind with a specific virus, bacterium, or protein (ELISA or blot tests).
- Genomics – in which you look for the genetic sequences of specific organisms (using PCR amplification or by binding to a microarray).
- Proteomics – in which you look for proteins specific to each pathogen.
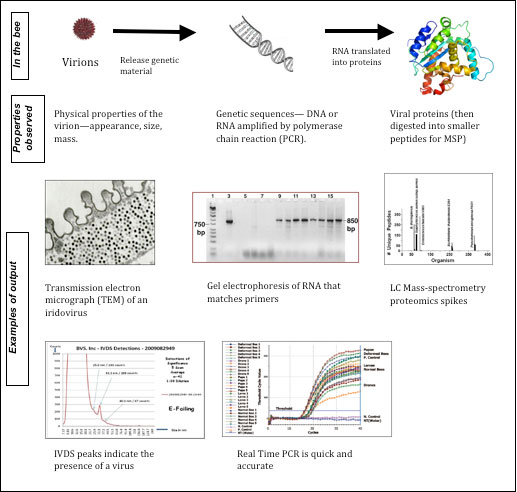
Figure 1. Different ways for identifying a virus. Please refer to my previous article about genetic transcription and translation. Each method has its advantages and disadvantages—price often being a consideration. (TEM and gel from Lapointe 2001; IVDS from Dave Wick; RT PCR from Chen 2005; MSP from Charles Wick; protein illustration from Wikipedia.)
Each of the above methods has its strong points and drawbacks when looking for known pathogens. But how about when you are trying to discover an unknown virus? It took twenty years to nail the HIV virus that causes AIDS; and believe me, lots of researchers were looking for it! Luckily, some of the newer technologies have made great strides—in 2002, Dr. Joe DeRisi’s “Virochip” identified the virus that was causing the SARS outbreak in less than 24 hours! (DeRisi, funded by Project Apis mellifera, is developing a microchip that should identify any bee pathogen in a sample.
It is much more difficult to identify an unknown pathogen. Realize that the study of bee pathogens has been greatly hampered by the lack of honey bee cell culture lines in which parasites could be isolated and grown. Luckily, an alternative host cell culture has recently been found for Nosema apis and ceranae, which may help greatly in the study of the pathology of these organisms (Gisder 2010); however, a true bee cell culture line is greatly needed, especially for viruses.
In the case of an unknown virus, the virions are so small, and in “covert” infections so sparse, that they may be entirely missed by electron microscopy. A novel virus may form a peak on IVDS, but very large viruses (such as IIV) may break up in the device. Dave Wick (pers comm) has shown me IVDS spikes that may indicate IIV.
Immunological techniques depend upon creating an exactly matching antibody, so unless the virus is closely related to the antibodies used, it won’t bind. Genomic methods depend upon creating the right “primers” (Fig. 2) that will bind to the viral RNA or DNA. DeRisi’s Virochip is imprinted with snippets of DNA from every virus ever discovered — about 22,000 different viral sequences—and with luck will bind to some part of any novel virus (however, his current chip did not find any iridovirus sequences in the Bromenshenk samples).
Figure 2. Typical published genetic primers. The four letters stand for the four base molecules that make up the backbone of DNA (e.g., G = guanine). Each “triplet” of bases codes for a specific amino acid. (Primers from Bourgeois (2010); codon graphic from Wikipedia.)
The key to creating such primers is to identify critical genetic sequences that are “conserved” as new viruses evolve from older viruses. Primers made from such “conserved regions” will likely hit on novel strains of that virus. Successful primers are generally posted to Genbank for use by all scientists. The problem with primers is that if you don’t get them exactly right, the mystery virus nucleic acids may not bind to them.
Proteomics differs from the above methods in that it looks at the final proteins that actually make up an organism (rather than the genes), and is useful for identifying traits in bees such as cold tolerance or parasite resistance. The process chemically digests all the proteins in a bee sample into smaller units called “peptides,” runs them through a liquid chromatography column to separate them by size, and then gives them an electrical charge and sprays them into a device that records the “spectrum” of their atomic masses (the mass spectrometer).
The data output is mind boggling: some 26 columns wide by thousands of rows deep for each sample! A computer then matches the peptide fragment data against the full library of known peptide sequences. The more peptides that match a catalogued pathogen’s “signature,” the more closely the unidentified pathogen is related to it. However, there is a drawback to the method with regard to identifying pathogens that may be present at low levels—their peptides may be lost in the confusion of all the other peptides in the sample (from both the bee and all its gut bacteria).
This is where Wick’s team had a brilliant idea—to combine proteomics with genomics! Still with me? Take a coffee break if necessary at this point, because understanding the next part is critical for appreciating the significance of the Army team’s whiz-bang new method.
A NOVEL APPROACH
What Wick’s team did was to download the genetic code for every bacterium, fungus, and virus (that had been fully sequenced as of September 2008). They then used a computer program to translate those codes into what would be the full complement of proteins for each organism, which were then “digested” by the computer into the expected peptides that would be seen in the mass spectrometer. At this point they had created a theoretical “mass spectrum” for each microorganism.
However, many peptides would be identical to those of other organisms, including the bee. So the team developed another computer program to discard any peptides that were not unique for each specific pathogen (all the bee and common pathogen peptides were “subtracted”). What was left was a “signature” of unique peptides for each microorganism. Their novel (patent pending) BACid computer program was then used to search for these microorganism signatures in any processed bee sample, and assign them by standard taxonomic classification, often right down to species and even strain (Figure 3).
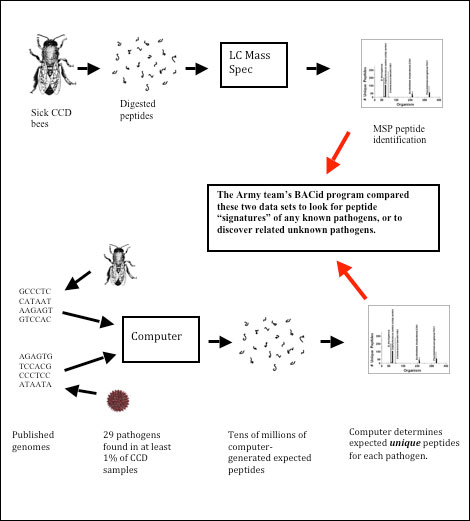
Figure 3. The upper path indicates how the Edgewood Lab processed CCD samples. The lower path is a schematic of how their novel computer program generated expected peptide sequences from the published genomes of bee pathogens, and then subtracted any peptides that were common to either healthy bees or to multiple organisms. This process then allowed them to look solely for the unique peptide “signature” of any specific pathogen. (Bee graphic from Kauffeld 1980).
The developers of the method suggested that it might “function as a strong complement to the alternative approaches of comparing microbial genomes based on DNA sequencing or microarray hybridization techniques” (Dworzanski 2006). And indeed it does, some major advantages of the method being:
- It is very rapid, and most of the work is done by computer.
- The approach allows for the detection, quantification, and classification of fungi, bacteria, and viruses in a single analytical pass.
- Classification can be to strain level and is limited only by the level of precision within the proteomic and genomic databases.
- It isn’t dependent upon the often tricky chemistry involved in standard genomic methods.
- Instead of using a limited number of genetic primers, it uses hundreds of confidently identified peptide sequences derived from the entire genomes.
- The signature isn’t likely to be misled by single point mutations, as such changes would likely affect peptides less than they would affect primers.
This technique appears to be a major breakthrough for pathogen identification. The Army’s pretty proud of it! As more bacteria, fungi, and viruses are sequenced each year, their proteomes can be added to the database, constantly improving the method. The method is explained in detail in a recent paper by the Edgewood team–Jabbour (2010).
Another of the technique’s beauties is that once you process a sample, the data set can be later “mined” should any new pathogens be discovered in the future—their peptide signatures will still be in the original data. Bromenshenk has been processing bee samples to save examples of the current “state of the bee” for posterity (researchers are acutely aware of the lack historical bee specimens suitable for parasite confirmation). Future researchers could then use the archived data to see how any new parasites change the dynamics of bee symbionts and pathogens, or to see whether a newly-identified virus was previously present, but merely unnoticed.
BACK TO THE PAPER
In the Results, the authors make an amazing claim:
“MSP analysis resulted in a database of more than 3,000 identifiable peptides, representing more than 900 different species of invertebrate-associated microbes.”
Nine hundred species of microbes? This is far more than all the bee symbionts and pathogens combined! The Army’s explanation, soon to be released in a technical report, is that bees are essentially flying dust mops, and pick up every imaginable microorganism in the environment, albeit at very low levels. The MSP analysis is simply so dang sensitive that it picked up, for example, traces of every organism being studied in the various labs at Bromenshenk’s university! MSP will even detect human skin keratin should you make the mistake of touching a bee with your bare finger when collecting the sample (Bromenshenk, pers comm).
“We narrowed the list of suspect microbes to those infecting bees and insects, 121 in all. Of these, only 29 were specific to bees or occurred in more than one percent of the colonies sampled…. Peptides were identified from nine of the approximately 20 known honey bee viruses.”
The authors narrowed down their analysis to known bee pathogens, plus included any organisms that were present in more than one hive out of a hundred. They did not waste time on well known bacterial pathogens such as AFB or EFB, which had already been ruled out since they would have been readily identified by CCD investigators.
A criticism of the paper that I’ve heard is that the MSP could have simply misidentified normal bee peptides as erroneous indicators of IIV. This suggestion does not hold water, in my humble opinion, for two reasons:
- The computer program (the algorithm) intentionally removed any bee peptides from consideration, and
- The team also sampled two control groups as a check—bees from Australian packages, and from an isolated, non-migratory operation in Montana that starts fresh with packages each spring. No IIV peptides were found in either of those groups, and only a single instance of a virus (Sacbrood) was found in the Montana hives. If the MSP was misidentifying bee peptides as something else, it should have done so in the control hives as well.
The meat of the paper is in Table 1, which shows the mean peptide count for various viruses and nosema in three groups of samples: from (1) bees from commercial apiaries sampled across the U.S. in 2006–2007, (2) bees sequentially sampled as the disorder progressed in an observation hive at Bromenshenk’s lab in 2008, and (3) bees from a recurrence of CCD in Florida in 2009.
Israeli Acute Paralysis Virus (IAPV) did not occur frequently, and was more prevalent in East Coast and Australian bees. This certainly surprised me, as it was found in my own West Coast hives two years ago, and I observed the devastating effect of inoculating colonies with IAPV in my Remebee trial.
“The most prevalent viral peptides we detected were identified as invertebrate iridescent viruses (IIV).”
This claim is the lightning rod for the paper—researchers who have not found indications of IIV in their samples are, to say the least, skeptical! At this point the MSP reached its limit, since it only had in its database two complete IIV sequences—that for IIV-6, which is a common insect pathogen, and for IIV-3, which has only been found in one species of mosquito. Until more types of IIV are genetically sequenced, the computer program cannot match the peptides to the actual strain of IIV apparently present. The authors explain:
“These procedures may have identified IIV-6 as the most likely source of peptides because this is the only fully sequenced genome from the genus Iridovirus. We suspect that bees may in fact be infected by IIV-24 that is also assigned to the Iridovirus genus, which was isolated from an Asian bee…, or by a variant of IIV-6.”
IIV-6 is a common insect virus which is easily cultured in wax moth larvae. I suppose that one could suspect contamination of samples from a bee brushing against an infected wax moth larva, but if you look at the average peptide counts (the in Table 1), that explanation stretches credulity, as no actual worms were in the samples. There would also be little reason for any IIV levels from wax worms to correlate with nosema.
By the way, this is a different virus than the Densovirus that has plagued the wax worm bait industry. However, I was recently viewing a YouTube video of author Dr. Shan Bilimoria, which showed sick wax worms infected with IIV-6. That image immediately jolted in my memory that I had noticed similar-looking sick wax worms when I first experienced colony collapse issues in 2005. It stuck in my mind because I had never seen sick wax worms before – they were always so danged healthy! So there is the possibility that a new strain of IIV has jumped hosts, from wax worms to bees (or vice versa; I’m keeping my eye open for the recurrence of sick wax worms).
The IIV-24 that they mention is also known as Apis Iridescent Virus, and can cause serious bee mortality in the Asian bee, Apis cerana. Bailey, Ball & Woods (1976) state that IIV-24 “multiplied when injected into adult individuals of Apis mellifera, and… multiplied abundantly when injected into young pupae. These developed more slowly than pupae injected with water, but some occasionally matured into seemingly normal adults containing much virus.”
So could a form of IIV-24 be the culprit? Bailey & Ball (1978) state that: “Apis iridescent virus was plentiful in each of several samples of adult individuals of Apis cerana from sick colonies in Kashmir and Northern India. Almost every bee, of those examined individually, was infected with the virus, which caused an easily detectable iridescence in the fat body and most other internal organs.” So if the putative IIV were indeed IIV-24, you’d think that we’d easily see the iridescent color (caused by diffraction of light by the densely-packed virions). So I’m guessing that it’s not IIV-24.
Just to make things interesting, I must note that vanEngelsdorp, et al (2006), when examining CCD bees under the ‘scope, state that: “Crystal-like formations were observed in the thorax where muscles are located. Similar structures have been described in some viral infections; however, it is not clear if these are the same type of structures.” Another odd thing is that IIV-24 is one of the few IIV’s that doesn’t appear to infect wax worms.
As long as we’re speculating, some IIV’s exhibit an orange-yellow color in heavily infected insects (Henderson 2000), which makes me wonder about the bright “corn-yellow” bee larvae that many of us are seeing in sick colonies (vanEngelsdorp, et al 2009). However, that infection appears to be controlled by treatment with oxytetracycline (personal experience), which does not support the virus hypothesis, plus iridoviruses normally do not replicate at normal broodnest temperatures. Likely just coincidence, but makes me curious.
There are also a couple of other pieces of evidence that an IIV might be involved in CCD. Dr. Mariano Higes, the Spanish N. ceranae researcher, found what appeared to be an IIV virion in a bee sample from a collapsing colony. Closer to home, in 1998, following unusual levels of winter mortality in the northeastern U.S., Camazine (1998) found “hexagonal, isometric particles resembling invertebrate iridoviruses” in the varroa mites! (But by the time he found them, it was too late to inspect the bees).
Since I’ve already crossed the speculation border, I’ve noticed that several CCD surveys have reported that mites were either not present, or at very low levels in collapsing colonies. The researchers generally assumed that the beekeeper had recently killed them with a miticide. But what if an iridovirus is killing the mites? Oh, this paper has sure opened a can of worms!
Still speculating, could varroa be the original or alternative host of the IIV? And further, could this be a jump of yet another parasite from Apis cerana to Apis mellifera, following varroa and N. ceranae, and perhaps tracheal mite, Kashmir bee virus, and who knows what else? Clearly, our bees appear to be finally picking up the entire suite of parasites from their cousins! And are those parasites, notably varroa, N. ceranae, IIV and perhaps some of the KBV/ABPV/IAPV group all co-adapted and interactive?
Q&A
I should pause for a moment at this point, and discuss some appropriate and legitimate questions about the purported identification of an iridovirus in Apis mellifera, and its linkage to CCD.
Q: How could these guys find this large virus, when no one else had seen it?
A: Others may have seen it, notably Higes and Camazine, but those were isolated incidents, and only tentatively identified by appearance. The paper suggests that: “inapparent infections by iridescent viruses may involve a low density of IIV particles in infected host cells, so without sensitive techniques such as MSP, it is not surprising that infections in CCD bee colonies were previously missed.”
The above answer is less than compelling to doubters, who are going to require additional confirmation before they will accept the finding as conclusive. One other point that must be kept in mind is that little analysis has been done of the “disappeared” bees (since they are so hard to find), so we really do not know the pathogen levels in the missing bees!
Q: How robust do they feel about their identification of IIV?
A: The Army does not go to press unless they think that they are incontrovertibly right (it’s a guy thing). The authors state:
“The large number of IIV proteins that we identified, 139 in all, represent a significant fraction of the total IIV proteome…belying any criticism that our identification of IIV may be a spurious consequence of accidental matching of a few peptide fragments.”
Q: How do they know that they are not simply misidentifying bee peptides as IIV peptides?
A: In the first place, their method generated the entire bee proteome, which they then “subtracted” from the results. Second, two “control” groups (Aussie bees, and “clean” Montana bees) exhibited no IIV peptides, thus supporting the authors’ contention that IIV peptides are not present in uninfected bees. It also suggests that IIV may be related to the presence of varroa, since no IIV was found in samples from the two bee groups that weren’t infested with varroa.
Q: Could IIV have become endogenized (incorporated into) into the bee genome, and the peptides thus have been generated by the bees themselves?
A: See above—no IIV peptides in the control groups. Also, the sheer number of IIV peptides observed would not be expected from expression of endogenized genes.
Q: Why didn’t the team show electron microscope images of IIV virions to validate their finding?
A: I put the question to Trevor Williams, the IIV expert. His answer:
“Hello Randy – it’s a good question; a picture paints a thousand words. Hopefully this issue will be resolved over the coming months and we’ll get the image you’re asking about. Finding virus particles in sublethally infected hosts can be extremely difficult – – maybe you remember how long it took for researchers to get a photo of HIV; this is because when the particles normally exist at a LOW DENSITY in covertly infected hosts, finding a cell with some particles inside is like looking for the proverbial needle in a haystack. Knowing which tissue to look at is an important step (one which we don’t have a good handle on in the case of the bee IIV).
“Only one person has managed to observe IIV in a sublethally infected insect– a student from the Czech Republic found a low density of virus particles in some gut cells from covertly infected mayflies– but this was a serendipitous finding.”
Bromenshenk’s team worked on a shoestring, and electron microscopy is pricey. They didn’t waste time on a likely futile search, since they weren’t seeing bees with blue tissues, but this is something that the team is currently actively pursuing.
Q: Why didn’t Bromenshenk get PCR or other genetic confirmation of the IIV?
A: Well, in a manner of speaking, they did! Their matches were based upon in silica (by computer) translation of all genetically sequenced IIV’s. As far as PCR, ELISA, Southern Blot, or microarray, they are all dependent upon creating a specific primer derived from sequencing of the virus, or a very close relative. Without the proper primer, you simply won’t detect the virus. Team member Dr. Robert Cramer is furiously working at sequencing the new IIV, collaborating with Williams and Bilimoria in his efforts. Indeed, he and Bilimoria and are currently (October) testing the efficacy of an antibody to IIV-6 as a diagnostic tool. If they are successful, then any of several rapid and relatively inexpensive tests for this specific strain of IIV could be made available.
Q: Why didn’t other scientists pick up IIV via genetic analysis?
A: This appears to me to be strongest scientific question of the putative IIV identification. Eaton (2007) found that all known members of the iridovirus family share quite a number of “conserved” genes. The genomists that I’ve spoken with feel strongly that they would have picked up the conserved genes. When this question is eventually answered, it will have great implications for the validity of the Army’s method vs. metagenomics (the method used by Cox-Foster/Lipkin in the 2007 paper that indicated that IAPV was a marker for CCD).
Q: IIV peptides were present in 9 out of 13 strong colonies; how do they explain that?
A: IIV peptide counts in strong colonies were less than half of what they were in failing colonies, and IIV was present in 100% of failing or collapsed colonies. IIV’s often exist in inapparent infections, in which the bees do not appear to be sick. Perhaps as long as nosema levels are low (or some other trigger is not present), IIV might not cause significant problems.
Q: Why did MSP indicate peptides from 10 species of nosema?
A: Because at the time of the analysis, Nosema ceranae had not yet been sequenced, so the computer matched to the closest nosemas. The Army plans to run the analysis again with the recently published genome sequence of N. ceranae.
Q: What the heck does the funny graph and weird statistics about “Discriminant Function Analysis” mean?
A: You’re asking the wrong person, but I’ll give it a shot. Discriminant function analysis (DFA) was used to determine which patterns of pathogen occurrence best discriminate between the groups Strong, Failing, and Collapsed in the 31 colonies from 2006/2007. In this case, failing and collapsed colonies tended to have high levels of IIV, plus nosema and Black Queen Cell Virus (long associated with N. apis; Bailey 1983); the levels of these parasites was about 80% predictive for collapse. The remaining 20% was predicted by the very low levels of Deformed Wing Virus (DWV) in failing colonies!
This result is surprising, since DWV has been strongly associated with colony collapse since shortly after the arrival of varroa. DWV titers generally go in lockstep with mite levels. Could it be that the high levels of IIV in failing colonies is killing varroa, so that they cannot transmit the virus? (that was more wild speculation). Or, could IIV itself suppress other viruses? (this appeared to be the case in Bromenshenk’s observation hive). Please realize that there were only a relative handful of colonies in each group in this analysis, so I’d be careful about extrapolating these results to all instances of CCD.
The authors had a chance to later test their discriminant functions on nine colonies from the 2009 Florida collapse event—three each of strong, failing, or weak. They performed pretty well at sorting the colonies: none were classified as “healthy,” and the rest were sorted fairly accurately based upon pathogen patterns.
Q: Was the virus the same in all sick colonies?
A: The authors finally looked at the similarity in occurrence of specific iridescent peptides in the three different groups of samples (2006/2007, obs. hive, Florida). The correlations seem surprisingly low to me if indeed a single IIV were involved in all the colonies sampled. However, the best correlation was between the 2007-8 East/West samples and the 2009 Florida collapse, which are likely the most representative data.
There is another aspect of IIV biology that may help to explain the diversity of IIV peptides. Williams (1998) observed: “Moreover, in all cases [of IIV infection] a marked degree of genetic heterogeneity [(differences)] was observed among the various [virus] isolates analyzed; identical isolates were never recovered from two different host larvae” (emphasis mine). I find this of great interest, since DNA viruses generally have much lower mutation rates than RNA viruses. The observation that the virus is slightly different in every single individual infected insect begs for further research.
Q: What else did they do to corroborate their conclusions?
A: In another small set of data, the authors tracked the counts of IIV and nosema peptides in samples of bees from Bromenshenk’s observation hive as it slowly collapsed over a period of five weeks—the counts were similar to those of failing colonies.
Finally, Cramer infected bees in cage trials with N. ceranae and a cultured strain of IIV-6 (he has yet to be able to isolate a pure strain of the IIV in their samples). I found the results to be supportive, but of limited significance. The purpose of such an experiment would be to fulfill Koch’s Hypothesis that inoculation of healthy colonies with the two pathogens will result in CCD-like collapses, but this will be impossible until they are able to isolate the virus.
Dr. Cramer tells me that it has been problematic for him to culture the virus, since other viruses are normally present, and especially since there is no available cell culture line for honey bees in which to grow it. However, he is optimistic about the progress that he is making, and will run similar cage trials as soon as he has a pure culture.
IRIDESCENT VIRUSES
Beekeepers outside of India or Pakistan have likely not heard of iridescent viruses prior to the Bromenshenk paper, so let’s start with a little background on them. Luckily, Dr. Trevor Williams maintains a great website, with plenty of free downloads (see References), from which I’ve obtained most of the following information.
IIV infections are common in invertebrates, but generally occur as covert infections, with no obvious signs of disease. They only occasionally flare up into epizootics, usually when host densities are great, and the weather is cool and damp.
Note that the characteristic colored iridescence in “patently infected” (dying) hosts is an unreliable indicator of infection, since low-level infections with no obvious signs of disease are much more common.
Little is known about how they actually cause disease in the host. However, these viruses have dramatic inhibitory effects upon the synthesis of DNA, RNA, and cellular proteins, and it is likely that they encode multiple proteins that suppress host immune responses. They also game another host immune response—they inhibit apoptosis (cellular death) to prevent the host from clearing the infection (by sacrificing infected cells), only to later induce apoptosis once the cell is packed full of newly-formed virions! IIV’s make our common bee viruses look like amateurs!
Another stunning observation is that: “Purely in terms of numbers, [IIV’s] are among the most efficient insect viruses in turning host resources into virus particles. Around 25% of the dry weight of a dead insect may be virus.” (This brings to question why CCD researchers haven’t found bees full of iridovirus).
The reasons that IIV infections occasionally progress from covert to patent is poorly understood. Williams (1998) explains that the viruses “may show different grades of virulence in different individuals.” Williams further observes that: “Certain [IIV’s] appear to exploit multiple hosts in their natural habitat” (like maybe varroa, wax moth, yellowjackets (which also harbor KBV), or the Small Hive Beetle?).
IIV’s have been tested as use for biocontrols, but have been found to have too low a rate of infectivity and slow speed of kill. However, Williams makes a chilling suggestion: “Perhaps the only real hope for the successful use of [IIV’s] in insect biocontrol lies in the development of formulations that augment the infectivity of the virus (e.g., by interacting with other pathogens)” (Nosema ceranae and varroa come to mind).
So what if the combination of IIV and N. ceranae is indeed what’s knocking out colonies?
Nosema: You can start by making sure that nosema doesn’t get out of hand. I’ve written extensively about N. ceranae, which is now the predominant species in the U.S. (Rose 2010). Fumagillin, properly applied by various methods, is a proven cure; other treatments have less data to back them up.
However, N. ceranae alone does not appear to always cause noticeable problems; unfortunately, there is as yet no cheap test to see whether one has IIV in their operation. However, the general biology of iridescent viruses suggests some practical implications:
Dampness: IIV’s require damp conditions to efficiently transmit. This fact bolsters the common advice to avoid placing apiaries in cool, shady, damp areas. Colonies always do better in warm, sunny locations. IIV virions degrade quickly in warm, dry conditions, so dry out deadout equipment before restocking.
Temperature: For some reason, IIV replication generally appears to be favored at about 70°F, and is completely halted at temperatures above about 85°F. Note that the bees in the insulating “shell” of the winter cluster drop to about 55°F, and that even the center of the cluster may drop to near 70°F if no brood is present. This suggests that you may wish to manage your bees to maintain strong colonies that can rear a bit of brood during the winter. The problem, though, appears to have more to do with unexpected cold snaps or lack of honey stores for heat generation, than with steady cold.
Disinfection: IIV’s are very sensitive to drying, UV light, and disinfectants (Nalçacıoğlu 2009). This may help to explain why CCD-affected beekeepers have observed that it is helpful to allow deadout combs to “rest” a while before restocking with bees.
Varroa: It appears that mites may vector IIV’s. Keep varroa levels down!
Broodless splits: The isolated Montana operation in the study was nearly mite- and virus-free. This was apparently due to them starting with fresh packages each spring. Starting up fresh yards each season with mite-treated, broodless packages or splits might be a good idea.
Supplemental Feeding: This may sound like a platitude, but those beekeepers who maintain protein levels by feeding pollen substitutes appear to have fewer problems than those who allow their colonies to become nutritionally stressed. This observation is supported by recent research which found that protein feeding helped caged bees to resist infection by a virus (DeGrandi-Hoffman 2010).
Bee/semen import protocols: Should MSP indeed prove to be able to detect pathogens at very low levels, it could be of great utility for screening potential breeding stock for import.
PRACTICAL APPLICATIONS
So what if the combination of IIV and N. ceranae is indeed what’s knocking out colonies?
Nosema: You can start by making sure that nosema doesn’t get out of hand. I’ve written extensively about N. ceranae, which is now the predominant species in the U.S. (Rose 2010). Fumagillin, properly applied by various methods, is a proven cure; other treatments have less data to back them up.
However, N. ceranae alone does not appear to always cause noticeable problems; unfortunately, there is as yet no cheap test to see whether one has IIV in their operation. However, the general biology of iridescent viruses suggests some practical implications:
Dampness: IIV’s require damp conditions to efficiently transmit. This fact bolsters the common advice to avoid placing apiaries in cool, shady, damp areas. Colonies always do better in warm, sunny locations. IIV virions degrade quickly in warm, dry conditions, so dry out deadout equipment before restocking.
Temperature: For some reason, IIV replication generally appears to be favored at about 70°F, and is completely halted at temperatures above about 85°F. Note that the bees in the insulating “shell” of the winter cluster drop to about 55°F, and that even the center of the cluster may drop to near 70°F if no brood is present. This suggests that you may wish to manage your bees to maintain strong colonies that can rear a bit of brood during the winter. The problem, though, appears to have more to do with unexpected cold snaps or lack of honey stores for heat generation, than with steady cold.
Disinfection: IIV’s are very sensitive to drying, UV light, and disinfectants (Nalçacıoğlu 2009). This may help to explain why CCD-affected beekeepers have observed that it is helpful to allow deadout combs to “rest” a while before restocking with bees.
Varroa: It appears that mites may vector IIV’s. Keep varroa levels down!
Broodless splits: The isolated Montana operation in the study was nearly mite- and virus-free. This was apparently due to them starting with fresh packages each spring. Starting up fresh yards each season with mite-treated, broodless packages or splits might be a good idea.
Supplemental Feeding: This may sound like a platitude, but those beekeepers who maintain protein levels by feeding pollen substitutes appear to have fewer problems than those who allow their colonies to become nutritionally stressed. This observation is supported by recent research which found that protein feeding helped caged bees to resist infection by a virus (DeGrandi-Hoffman 2010).
Bee/semen import protocols: Should MSP indeed prove to be able to detect pathogens at very low levels, it could be of great utility for screening potential breeding stock for import.
DISCUSSION
The conclusions of this paper have generated intense questioning and skepticism, which was exacerbated by the press hoopla about the mystery of CCD being “solved,” which I feel is inappropriate at this point, since (replication of the identification aside) there have not yet been any infectivity studies.
The main sticking point is that the discovery is based upon “deep mining” of machine- and computer-generated data. The Army method searched for correlations between the masses of ionized peptide fragments, and computer-generated hypothetical peptide sequences—no one actually saw or physically confirmed the presence of the iridovirus. I can guarantee that the findings will be controversial until one of the above is done!
Putting doubts aside, let’s assume that the team is correct, and discuss the implications of their findings. What I can say is that the results support what appears to be shaping up as a generic model for honey bee problems worldwide, both historically and in the present:
One or more viruses + nosema or varroa + chill events = sick colonies
This should come as no surprise to you if you’ve read the generic model for colony collapse that I published in the September issue of this journal. Allow me to quote from the CCD Action Plan: “Even if CCD is cyclic, it could be caused by a different pathogen in each case; for instance, a new pathogen could be causing significant bee loss (CCD) until the bees are able to develop resistance, at which point the problem disappears until the emergence of the next new pathogen.”
Things have gone seriously downhill for beekeepers (of Apis mellifera) in all countries in which varroa and Nosema ceranae have been introduced. These two parasites appear to have rewritten the rules of virus dynamics and general colony health.
If indeed there is a previously overlooked iridovirus involved, it may help to explain some vexing questions, such as why N. ceranae seems sometimes to be so devastating, yet at other times relatively benign.
From a scientific bent, perhaps the most significant aspect of this paper is that it was the first field test of a revolutionary pathogen discovery method developed by scientists working for the U.S. Army. If the results are validated, it will open a new era of pathogen forensics, allowing scientists to step out of the light of the figurative streetlamp, and search more effectively in the darkness.
For this, beekeepers owe the Army a great debt of gratitude for all the free work that they’ve done in our behalf. Will the Army’s findings be vindicated? We will find out soon enough, but I can say, the Army doesn’t like to be embarrassed, so these findings would not have been released lightly.
Of considerable import is that should the team’s purported discovery of a new virus in CCD colonies be confirmed, it would then call into question virtually all previous CCD research, since there is no way of knowing whether the purported IIV was present in the previous studies, unless the samples are reanalyzed! This is similar to the case of any Nosema apis research performed over the past decade or so—now that we’ve learned that N. ceranae invaded a number of countries unnoticed during that time. We must now question which nosema the researchers were actually working with!
Is the putative unidentified iridovirus the cause of CCD? Author Dr. Shan Bilimoria explains: “At this stage, the study is showing an association of death rates of the bees with the virus and fungus present. Our contribution to this study confirms association. But even that doesn’t prove cause and effect. Not just yet” (Davis 2010).
The team is pursuing funding for the logical next steps. Bromenshenk explains: “We have a proposal pending to isolate, characterize, and then inoculate bees with the specific iridescent virus that occurs in USA bees. This is a critical step, since the virus does not appear to be any of the world’s known iridescent viruses. Once we have the actual virus, we can complete the inoculation trials that are needed to test whether we’ve truly found the cause of CCD.”
Meanwhile, the internet is awash in “gotcha” blogs about the evil Dr. Bromenshenk. Writers who couldn’t tell a beehive from a birdhouse are fueling a feeding frenzy for conspiracy buffs (for more rational views, see Blogs).
So will Bromenshenk and team suffer the fate of Stanley Pons, who stunned the world in 1989 with the (subsequently unreproducible) claim that he had achieved cold fusion, or will the team be vindicated by subsequent research, thus forcing their detractors to seek recipes for eating crow? Only time will tell.
ACKNOWLEDGEMENTS
As always, I must express my heartfelt gratitude to my co-researcher Peter Loring Borst. I also appreciate the time given me by the various authors of “The Paper,” as well as by some of its not to be named critics.
REFERENCES
The Paper
Bromenshenk JJ, Henderson CB, Wick CH, Stanford MF, Zulich AW, et al. (2010) Iridovirus and Microsporidian Linked to Honey Bee Colony Decline. PLoS ONE 5(10): e13181. doi:10.1371/journal.pone.0013181. Free download at http://www.plosone.org/article/info:doi/10.1371/journal.pone.0013181#abstract0
Mass Spec Proteomics (LC-MS-MS)
The most appropriate paper for describing the Army’s method and accuracy is:
Jabbour RE, et al (2010) Double-Blind Characterization of Non-Genome-Sequenced Bacteria by Mass Spectrometry-Based Proteomics. Applied and Environmental Microbiology 76(11): 3637-3644. http://aem.asm.org/cgi/content/abstract/76/11/3637 Unfortunately not a free download.
MSP is well reviewed by Shushan in the two following papers:
Zonderman, J, and B Shushan (2008) Liquid Chromatography Coupled with Tandem Mass Spectrometry for Clinical Applications. Free download at (Broken Link!) http://spectroscopyonline.findanalytichem.com/spectroscopy/Mass+Spectrometry/Liquid-Chromatography-Coupled-with-Tandem-Mass-Spe/ArticleStandard/Article/detail/566989
Shushan, B (2010) A review of clinical diagnostic applications of liquid chromatography–tandem mass spectrometry. Mass Spectrometry Reviews, n/a. doi: 10.1002/mas.20295.
Wick, C (2009) Unrestricted Biological Detection Methods http://www.nanotecnexus.org/files/charles-wicks-presentation—environmental-application.pdf
Jabbour, et al ( 2010 ) Identification of Yersinia pestis and Escherichia coli Strains by Whole Cell and Outer Membrane Protein Extracts with Mass Spectrometry-Based Proteomics. Journal of Proteome Research 9: 3647–3655.
Iridoviruses
Plenty of free downloads at Trevor William’s website: (Broken Link!) http://www.trevorwilliams.info/Iridovirus.htm
Williams, T. (2008) Iridoviruses of invertebrates. In: Encyclopedia of Virology (Third edition). B. W. J. Mahy & M. H. V. Van Regenmortel (Eds.), pp. 161-167, Elsevier, Oxford, UK.
Williams, T. (1998) Invertebrate iridescent viruses. In: The Insect Viruses (Eds. Miller L. & Ball A.) pp. 31-68, Plenum Press, NY.
Nalçacıoğlu, R, et al (2009) The Biology of Chilo Iridescent Virus Virologica Sinica 24 (4):285-294
Blogs
http://myrmecos.net/2010/10/13/honey-bees-as-pawns/
(Broken Link!) https://sharepoint.cahnrs.wsu.edu/blogs/urbanhort/archive/2010/10/13/ignorance-and-the-so-called-%E2%80%9Cbogus%E2%80%9D-bee-study.aspx
Citations
Bailey L, Ball BV (1978) Apis iridescent virus and “clustering disease” of Apis cerana. J Invertebr Pathol 31: 368–371.
Bailey, L, BV Ball, and RD Woods (1976) An Iridovirus from Bees. Journal of General Virology 31: 459-461.
Bailey, L, BV Ball, and JN Perry (1983) Association of viruses with two protozoal pathogens of the honey bee. Ann. Appl. Biol. 103: 13-20
Billimoria, S (2009) Use of iridoptin to induce toxicity in insects. United States Patent Application US2009/0069239.
Bourgeois, AL,*, TE Rinderer, LD Beaman, RG Danka (2010) Genetic detection and quantification of Nosema apis and N. ceranae in the honey bee. Journal of Invertebrate Pathology 103: 53–58.
Camazine S, and TP Liu (1998) A putative iridovirus from the honey bee mite, Varroa jacobsoni Oudemans. J Invertebr Pathol 71: 177–178..
Chen, YP, J. A. Higgins,2 and M. F. Feldlaufer (2005) Quantitative Real-Time Reverse transcription-PCR Analysis of Deformed Wing Virus Infection in the Honeybee (Apis mellifera L.). Applied and Env. Microbiology 71(1): 436–441.
Chinchar, V.G., Hyatt, A., Miyazaki, T. and T Williams (2009) Iridoviridae: poor viral relations no longer. Current Topics in Microbiology and Immunology 328: 123-170.
Cox-Foster, DL, et al. (2007) A metagenomic survey of microbes in honey bee colony collapse disorder. Science 318: 283-287.
Davis, J (2010) Researcher: Bee Colony Collapse Associated with Viral, Fungal Infection. http://today.ttu.edu/2010/10/researcher-bee-colony-collapse-associated-with-viral-fungal-infection/
DeGrandi-Hoffman, G, Y Chen, E Huang, MH Huang (2010) The effect of diet on protein concentration, hypopharyngeal gland development and virus load in worker honey bees (Apis mellifera L.). Journal of Insect Physiology 56: 1184–1191.
Dworzanski JP, Snyder AP, Chen R, Zhang H, Wishart D, Li L. (2004) Identification of bacteria using tandem mass spectrometry combined with a proteome database and statistical scoring. Anal Chem.76(8): 2355-66.
Dworzanski,JP, SV Deshpande, R Chen, RE Jabbour, AP Snyder, CH Wick,and L Li (2006) Mass Spectrometry-Based Proteomics Combined with Bioinformatic Tools for Bacterial Classification. Journal of Proteome Research 5: 76-87.
Eaton, HE, et al (2007) Comparative genomic analysis of the family Iridoviridae: re-annotating and defining the core set of iridovirus genes. Virology Journal 4:11 http://www.virologyj.com/content/4/1/11
Evans, JD (2010) Microbes Associated with Honey Bee Colony Collapse. Presentation to the 10th General Meeting of the American Society for Microbiology in San Diego.
Gisder, S, N Möckel, A Linde and Elke Genersch (2010) A cell culture model for Nosema ceranae and Nosema apis allows new insights into the life cycle of these important honey bee-pathogenic microsporidia. Environmental Microbiology (in press).
Hackett, K, et al (2007) Colony Collapse Disorder Action Plan. http://www.ars.usda.gov/is/br/ccd/ccd_actionplan.pdf
Henderson, C (2000) Replication and pathogenesis of an iridescent virus in the cotton boll weevil. PhD Dissertation, , Texas Tech University.
Kauffeld, NM (1980) Beekeeping in the United States. Agricultural Handbook Number 335.
Lapointe, SL, Wayne B. Hunter, and C. J. Funk (2001) Infection of the Diaprepes Root Weevil, Diaprepes abbreviates (Coleoptera: Curculionidae) by an Iridovirus. (Broken Link!) http://www.ars-grin.gov/ars/SoAtlantic/fp/sti/lapointe/drw_iridovirus/
Rose, R, et al (2010) The National Honey Bee Disease and Pest Survey: 2009-2010 Pilot Study Summary Report. http://www.aphis.usda.gov/plant_health/plant_pest_info/honey_bees/downloads/2009_Pilot_Summary_Report.pdf
vanEngelsdorp D, Evans JD, Saegerman C, Mullin C, Haubruge E, et al. (2009) Colony Collapse Disorder: A Descriptive Study. PLoS ONE 4(8): e6481. doi:10.1371/journal.pone.0006481
vanEngelsdorp, D, et al (2006) Fall Dwindle Disease: A preliminary report http://maarec.cas.psu.edu/pressReleases/FallDwindleUpdate0107.pdf




