Selective Breeding for Mite Resistance, Part 3; Shifting the Genetics of a Breeding Population
Selective Breeding for Mite Resistance, Part 3
Shifting the Genetics of a Breeding Population
Randy Oliver
ScientificBeekeeping.com
The alleles necessary for varroa resistance already existed in my stock of bees, so I didn’t need to “create” anything new. What I’m attempting to do via strong selective pressure is to (1) eliminate from our breeding stock the alleles that favor varroa, (2) increase the prevalence of allelic combinations that confer mite resistance, and (3) “fix” those alleles in the genome of our breeding population. During this process, I want to (4) minimize any deleterious “bottlenecking” of our stock’s genetics. So let’s talk about those genetics.
Several important terms
Breeding population: a population within which free interbreeding takes place and evolutionary change may appear and be preserved.
Genome: The entire genetic structure of an individual or breeding population. All the individuals have the same genes, but the breeding population may contain a number of different forms (alleles) of any gene. In this case, we’re selecting for combinations of alleles that work together to confer mite resistance.
Genotype vs phenotype: An organism’s genotype is the set of alleles that it carries. An organism’s phenotype is all of its observable characteristics — which are influenced both by its genotype and by the environment. We’re selectively breeding for the phenotype of exhibiting very low mite counts; we can only guess at the genetics involved.
Fixation: In population genetics, fixation is the change in a breeding population so that only one allele of a specific gene remains in that population (such as the recessive allele that results in the cordovan phenotype).
Genetic bottlenecking: A reduction of allelic diversity in a breeding population. In the case of our selective breeding, we are intentionally trying to capture and bottleneck the diversity of the alleles associated with the trait of mite resistance (favoring the alleles that confer resistance), but want to conserve the diversity of the rest of genome.
Recurrent selection: A cyclic selection process that is used to increase the frequency of desirable alleles for a character or trait that already exists within that breeding population at low levels. The method involves repeated selection of, and breeding from, generation after generation of individuals exhibiting the desired trait. The interbreeding of their offspring then allows for novel genetic recombinations to occur, and ideally to eventually become “fixed” in that breeding population, without excessive reduction of overall genetic diversity within that population.
Practical application: We’re using the tried and true method of recurrent selection — breeding only from the queens of colonies that exhibit resistance to varroa — in the hope that we can fix the genetic recombinations that confer mite resistance into the genome of our entire breeding population.
The drone pool and avoidance of excessive inbreeding
Selective breeding of honey bees requires the genetic management of an entire breeding population, in this case, the thousand-plus colonies of our own stock dedicated to our program (since we replace every queen in our operation each year with one from our selected breeders). We bring our colonies back from almond pollination chock-full of drones, and flood our mating yards with the sons of last-season’s queens. We also provide free queen cells of our stock to nearby hobby beekeepers. Based upon research by Hellmich [[1]], I have reason to believe that we control most of the genetics of our drone pool.
Practical application: The 50+ bee yards in our operation are largely in wooded areas (which provide suitable cavities for feral honey bees). And although we largely manage swarming, in some years swarms do fly off and establish feral colonies. I do not know how much genetic difference there is between our local ferals and our managed stock, but the influence of our drones upon feral genetics would likely be considerable.
Any of our swarms that successfully establish, but don’t carry the alleles for strong mite resistance, will become feral “mite factories” in late summer. My own yet-unpublished data suggest that they account for much of the mite immigration coming into our managed hives. The good news is that non-resistant feral colonies will likely perish during the winter, and won’t be sending out drones the next spring, meaning that any drones coming from the feral population will likely carry alleles for resistance.
Another poorly-understood variable observed by Couvillon [[2]], is that drones from some mothers may be disproportionally more successful at actually inseminating a queen. Whether that has any linkage to the alleles for mite resistance is unknown.
Selection vs maintaining Diversity
It’s also important to maintain a large enough diversity of queen mothers each generation. Otherwise, excessive bottlenecking of the genetic diversity of your breeding population will catch up with you, resulting in an inadequate number of sex alleles in the population, and queens laying diploid drones (as well as lack of general genetic diversity to allow for environmental adaptation, and to mask the effects of deleterious recessive alleles). As explained by King [[3]]:
Non-random, imbalanced mating designs in which better parents are crossed more often does increase the per generational gains by nearly 10%. However, there is a cost associated in terms of reduced effective population size.
Practical application: In our breeding program, we graft off a minimum of 30 queens (all having mothered a mite-resistant colony) every year in order to maintain an adequate amount of genetic diversity (remember that every queen carries the genetics of dozens of drones) (Figure 1). I keep an eye on both brood patterns, as well as the wide variation in coloration in our queens as indicators of diversity.

Fig. 1 We maintain any colonies marked as “potential breeders” (due to having very low mite counts) in our outyards along with the rest of our colonies, subject to plenty of mite drift. Only after almond pollination do we move the 30-40 “best of the best” to our home yard (shown above) to provide larvae for grafting. To make “breeder grade,” a mite-resistant colony must come back from almonds chock-full of bees and almond honey. To prevent them from swarming, we reduce them to small singles, and only after grafting add back a second brood chamber back to allow them to regrow.
Trying to figure out the genetics involved
It’s important to understand that the queen is not directly involved in varroa resistance — it is the single or combined responses of the workers of the colony that affect the mites. Keep in mind that a colony consists of as many “patrilines” of full-sisters (also called “supersisters”) as the number of drones that the queen mated with — each patriline potentially exhibiting differences in behavior.
It could be that it takes only a single patriline of daughters to initiate varroa-sensitive uncapping behavior. And that behavior could be due to that patriline of workers inheriting a single dominant allele from their father.
Practical application: I’ve seen over and again that there can indeed be strong, gentle, and productive colonies that are absolutely bulletproof to varroa. But the queen’s genetics may have little to do with the resistance exhibited by the colony, due to the genetic influences of the different drones with which she mated. Thus, unlike the genetic heritability of a trait carried by the mating of a single mother with a single father, a trait exhibited at the colony level may be the result of a large number of different paternal bloodlines of workers all interacting with each other. This means that duplication of the genetics of a resistant colony is nearly impossible with open-mated queens, since there were so many fathers involved — any one of which could have been a rare “wild card.”
Allow me show you a couple of examples…
“Old School” Mendelian genetics
We’ve all heard of Mendelian genetics, and how to figure out the expected results from crosses of the genetics of two parents by using a grid of Punnett squares. Let me give the classic example for pod coloration of pea plants, involving a dominant allele (G) for green color, and a recessive allele (g) for yellow pods (Figure 2).
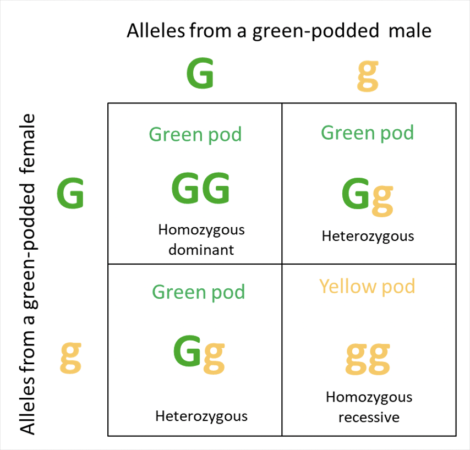
Fig. 2 One of Mendel’s Punnett squares, showing the results a test cross between two pea plants heterozygous for pod coloration, with coloration being determined by the alleles of a single gene. Note that the effect of the dominant allele (G), outweighs that of the recessive allele (g), so that the phenotypes of the parents (the green pods that we see) do not reflect their heterozygous genotypes. Caveat: in light of current knowledge, I’m not sure that pod coloration is actually this simple.
It gets far more complicated with honey bees, because the phenotype of the colony (such as resistance to varroa) may involve the genetics of a large number of parents. A diploid honey bee queen mates with dozens of haploid drones. Each drone thus fathers two patrilines of workers — one carrying the alleles of the drone combined with the alleles of the queen’s father, the other a combination with those of the queen’s mother [[4]]. I’ve illustrated this in the Punnett squares of Figure 3.
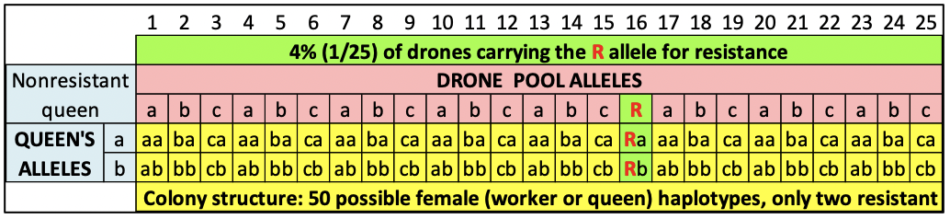
Fig. 3 Let’s hypothetically assume that there is a gene for uncapping behavior with several alleles — a, b, and c being common, but recessive and not triggering uncapping. But there’s also a rare allele R (for resistance) which is dominant, and does induce uncapping behavior. If a non-resistant queen mates with 25 drones, with only one carrying the R allele, this will create a colony consisting of 50 daughter patrilines (the yellow cells), of which only two (the green cells) will exhibit the behavior to uncap cells containing a mite — which may be able to confer resistance to the entire colony as a whole.
Practical application: The colony containing the 50 different genetic combinations of workers shown above might exhibit mite resistance, due to the performance of only the two patrilines of daughters from a single drone. But the queen herself would not carry any alleles for resistance.
If I were to graft daughters from the queen of the resistant colony above, only two out of 50 daughters would carry the R allele for resistance in their maternal bloodline; although some of the rest might be able to exhibit resistance at the colony level by the lucky chance of mating with an unrelated drone that did carry the allele.
Practical application: A great deal of luck is thus involved in picking the right daughters, since we don’t know whether the colony’s alleles for resistance came from the queen, or from one or more drones. It would be easy if the critical alleles were recessive, since the queen of any resistant colony would have needed to have carried that allele, and thus conferred it to half her daughters. But it’s much more difficult to pick a queen actually carrying a dominant allele, since any of the other 48 daughters might head a resistant colony if she happens to mate with a drone carrying that dominant allele.
In the case of selecting for a dominant allele, one must perform “progeny testing” — tracking the performance of the colonies of daughter queens, looking for queen mothers for which half of their daughter colonies exhibit resistance (Figure x), and then going back and breeding off them or the original mother. Last year was the first time that we saw this happen. We’ve got those resistant daughter colonies identified, and plan to move them to isolated mating yards this summer to attempt to “fix” the winning genetics into maternal bloodlines. Keep your fingers crossed!
Figure 4 shows the expected results of what we’d expect to occur if we got lucky (2 chances out of 50) and happened to graft from either of the two daughters above that carried the R allele in their maternal bloodline. In this case, not only would half the workers in her colony carry the R allele (and the colony likely exhibit strong resistance), but half of her daughter colonies would also be expected to exhibit resistance.
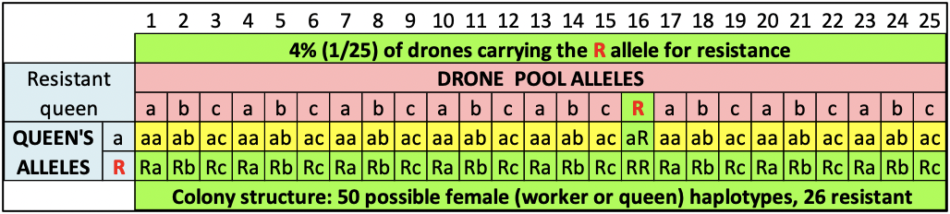
Fig. 4 An example of progeny testing. Same simulation as above, but this time with a queen carrying the R allele for resistance. In this case, fully half of that queen’s daughter colonies would now carry the critical R allele in their maternal bloodline.
Practical application: In one of our yards last season, 24 out of 48 colonies exhibited resistance. We’re crossing our fingers that we may have hit the jackpot!
I’ve spent hundreds of hours these past months running huge Punnett square series (up to 50 x 50 grids) to figure out the genetics involved in order to match our actual progress (shown in my previous article). I’ve run simulations with single alleles, double alleles, dominant or recessive alleles, epistatic dominant or recessive effects, etc. But every simulation suggests that even if the trait came originally from the rare drone, that by breeding only from mothers whose colonies demonstrate full resistance, we “should” be seeing more rapid progress.
Practical application: It’s relatively straightforward to select against a dominant allele with an observable phenotype, by culling individuals expressing that trait (e.g., cull the queens of any “hot” colonies). And it’s pretty straightforward to select for a recessive allele, by breeding only from individuals expressing that trait (e.g., breed only from cordovan-colored queens).
On the other hand, it’s difficult to select for a dominant allele (meaning against other recessive alleles), since you can’t tell whether a queen is carrying a recessive allele unless she’s homozygous for the recessive allele (refer to the pea illustration). See [[5]] for some good visuals of what I’m talking about.
And in the case of the “trait” of varroa resistance, although we might phenotypically observe uncapping behavior, the only proof that it actually conferred resistance to the colony would be to take mite wash counts.
Another reason that the genetics involved are so hard to understand is that the expression of particular traits is far more complicated than simple Mendelian genetics and Punnett square combinations. As explained by Miko [[6]]:
The relationship of genotype to phenotype is rarely as simple as the dominant and recessive patterns described by Mendel… Mendel’s early work with pea plants provided the foundational knowledge for genetics, but Mendel’s simple example of two alleles, one dominant and one recessive, for a given gene is a rarity. In fact, dominance and recessiveness are not actually allelic properties. Rather, they are effects that can only be measured in relation to the effects of other alleles at the same locus. Furthermore, dominance may change according to the level of organization of the phenotype. Variations of dominance highlight the complexity of understanding genetic influences on phenotypes.
Miko goes on to explain the concepts of partial dominance, co-dominance, overdominance, multiple alleles and dominance series. That said, in the simplest cases, one may be able to select for or against some easily-identifiable dominant or recessive traits, such as color, “gentleness,” or hygienic behavior. But mite resistance doesn’t appear to be so simple.
Practical application: The bottom line is that selective breeding for mite resistance is a challenge, especially since there are dozens of fathers involved in any resistant colony. I’ve given up on trying to understand all the complex genetics (and epigenetics) involved in mite resistance. So we just keep plugging away with our “recurrent selection” approach — which has a long history of success. In short, we simply requeen our entire operation each year solely with daughters of queens whose colonies exhibited control of varroa for the entire previous year.
What can small-scale beekeepers Realistically hope to do?
I often get asked by small-scale beekeepers how they could engage in a breeding program of their own. Unless they plan to instrumentally inseminate each of their queens, the question then is to what extent can they realistically expect to control the genetics of their drone pool?
Practical application: Honey bees may be the most difficult animal on Earth to selectively breed, due to the uncontrolled polyandry of the queens with the drones of the surrounding breeding population. The selective breeding experiment that my sons and I are engaged in is targeted for large-scale queen producers who run enough colonies to manage the genetics of an entire breeding population.
As you can see from our results, in which we essentially control the genetics of the drone pool, but also apply extreme selective pressure (which requires a lot of colonies to pick from), that it takes time and dedication.
The reality is that it’s difficult to imagine that any hobby beekeeper (unless they live on an isolated island) could be expected to significantly affect the breeding population surrounding their apiaries.
That said, if they are surrounded by an unmanaged population of native or feral honey bees, upon which Mother Nature herself has impartially applied enough selective pressure (“live and let die”) upon the free-living breeding population to gain resistance to varroa, the beekeeper could work in collaboration with the “natural” evolutionary process (nicely discussed by van Alphen [[7]]).
They could do so by eschewing bringing in non-resistant commercial stock, and instead populating their hives with swarms from the surrounding “wild” population. The beekeeper could then retain only colonies that were amenable for beekeeping (such as being gentle and productive), cull the queens of those that were not, and rear replacement queens from the “good ones.” I get reports from beekeepers in areas with viable unmanaged honey bee populations of doing just that, and being happy with the results.
Yes, such a beekeeper could consider themselves to be a “breeder,” but most of the credit would actually go to Mother Nature, with the beekeeper simply culling out bloodlines from their few managed colonies that were too spicy or swarmy. Nothing wrong with that!
Acknowledgements
I want to express my appreciation to my intellectually-gifted beekeeping friend Richard Cryberg, who has for years been an invaluable resource to answer my deep questions on chemistry, pesticides, biology, disease, and especially animal breeding and genetics. His deep knowledge and experience in the breeding of racing pigeons has helped me immensely in my experiment to selectively breed for varroa-resistant honey bees.
References
[1] Hellmich, R & G Weller (1990) Preparing for Africanized honey bees: Evaluating control in mating apiaries. ABJ 130(8): 537-542.
Hellmich, R, et al (1993) Evaluating mating control of honey bee queens in an Africanized area of Guatemala. ABJ March 207-211.
[2] Couvillon, MJ, et al (2010) Sexual selection in honey bees: colony variation and the importance of size in male mating success. Behavioral Ecology, 21(3): 520-525.
[3] King, J & G Johnson (1993) Monte Carlo simulation models of breeding-population advancement. Silvae Genetica 42(2-3): 68-78.
[4] Recombination and crossover of chromosomes during the process of meiosis complicates this even further — in reality every single worker bee (or potential queen) could theoretically be genetically unique.
[5] Selection for qualitative phenotypes. https://www.fao.org/3/v8720e/V8720E03.htm
[6] Miko, I (2008) Genetic dominance: genotype-phenotype relationships. Nature Education 1(1):140.
[7] Van Alphen, J & B Fernhout (2020) Natural selection, selective breeding, and the evolution of resistance of honeybees (Apis mellifera) against Varroa. Zoological letters 6(1): 1-20.



