What’s Happening To The Bees? – Part 4: The Genetic Consequences of Domestication
May 26, 2014
CONTENTS
So How Does This Apply To Bees?
Size And Survivability Of The Breeding Population
The Consequences Of Bottlenecks
So How Much Genetic Diversity Have We Lost?
What’s Happening to the Bees?
Part 4: The Genetic Consequences of Domestication
Randy Oliver
ScientificBeekeeping.com
Originally published in ABJ May 2014
I’m beginning this article at the point in time when beekeepers first learned to select for more manageable and productive bees. At the moment that humans began controlling the reproduction of honey bee stocks, the process of domestication was begun. This process has intrinsic genetic and biological consequences, some of which have come to haunt us today.
The Domestication Of The Bee
As humankind comes to depend more and more upon domesticated species to feed our growing population, animal and plant breeders are realizing the value of the genetic diversity of the wild stocks from which our domesticated species were derived. Chaudhary [1] explains that:
The term “domestication” is often used to describe the process by which wild becomes stabilized… domesticated forms are by definition wild species with certain traits highlighted under human selection.
He also wondered:
Is such…transformation under domestication universally advantageous or [is it] accompanied with the loss of an “additional” benefit? What is the spectrum of consequences of having a set of important genomic loci selected under human selection?
This is an exceedingly difficult question to answer, in large part because “success” is an ill-defined term that can refer to anything from short-term proliferation of individuals to long-term effects on lineage diversification.
Referring to the domestication of crop plants, he warns that:
Recent large-scale microarray studies on the comparison of wild and domesticated forms of selective plant species confirmed that global gene expression had been radically altered by domestication…Using DNA technologies, the diversity of domesticated tomatoes is estimated to comprise [less than] 5% of the genetic variation as compared to the rich reservoir in wild relatives.
Though the successful application of breeding programs has produced high-yielding crop varieties, ironically the plant breeding processes have threatened the genetic basis upon which the breeding depends.
So How DoesThis Apply To Bees?
In recent years, I’ve heard beekeepers complain again and again as to how our bees aren’t as “tough” as they used to be. A large part of this clearly has to do with the negative impacts of the varroa/virus complex, Nosema ceranae, and miticide contamination of our combs. But some recent visits to Southern California beekeepers made me suspect that there has been a more constitutional change in our bees in the aftermath of the varroa invasion.
SoCal beekeepers Rob Stone and Sean Crowley have generously taken time to allow me to inspect apiaries stocked with various combinations of commercial domesticated bees, local feral (partially Africanized) bees, or hybrids of the two—at the most stressful time of the season. I could not have been more impressed by the obvious differences between the races! Although I am familiar with the generally excellent performance of purchased bees under good management practices, the health and vigor of the feral bees under less than optimal conditions and management was striking. Although the hived ferals clearly exhibited “touchier” temperament, and focused their efforts more upon brood production than upon storing honey, I am haunted by the images of how fit and robust they were (Fig. 1), compared to the floundering domesticated stocks.
Figure 1. Sean Crowley holding brood frames from a typical Southern California hived feral colony (untreated for mites). This photo was taken on Sept. 27, at the end of the summer drought, shortly after a light fall nectar flow had begun. Nearby colonies of commercial stock at the same time had minimal brood, and were being eaten alive by varroa.
Those feral colonies reminded me of the bees of yore, which could be hived and then left to fend for themselves for years at a time, compared to the commercial bees of today, which are unable to survive without being coddled, fed, medicated, treated, and requeened on a regular basis. The question that occurs to me is, have our domesticated bees lost some of the innate vigor of their parental stocks?
Practical application: Two burning questions keep bugging me:
- Why are our bees such wimps compared to some feral stocks? And,
- why have the bees in their European homeland not yet evolved resistance to varroa, when some other races quickly developed resistance?
Although I may be taking a circuitous route to get there, please keep in mind that these articles are simply my semi-organized and evolving notes to myself as I try to answer the above two questions to my satisfaction.
The Cost of Domestication
In the Mediterranean and Central European environments modified by human colonization, the reproductive success of the honey bee was put largely under the control of beekeepers, by virtue of their controlling the supply of nesting cavities. And that changed the process of evolutionary selection for “fitness” from the ability of a colony to thrive in a natural setting, to its amenability to live as an animal domesticated by Man. “Fitness” now meant being gentle, productive, willingness to nest in manmade hives at ground level, and the ability to survive in close proximity to many other such hives.
Early in the domestication process, the selective process would have been little different from that exerted by Mother Nature, other than that humans would exert a strong negative selection against excessive colony defensive behavior (stinging). So at this point in time, the “semi-domesticated” bee population and the wild bee population would have freely interbred wherever there existed natural bee trees nearby. Later on, the selective processes for those two populations would diverge, a subject to which we will later return.
I was curious as to whether there is a price paid by a species (whether animal or plant) in the process of becoming domesticated, so I read up on the subject, and found it both fascinating and likely applicable to the domestication of the honey bee. Returning to Chaudhary, he asks:
What is the spectrum of consequences [in] genes and mutations underlying domestication transitions (colloquially called [the] “domestication syndrome”)?
Selection by humans for what we consider to be desirable traits may come at a cost in fitness of that domesticated breed should it be forced to face the stresses of nature without human support. As a bee breeder, I find this subject to be worthy of deeper investigation. As a biologist, I’ll start by going straight to the heart of the matter.
Genotype vs. Phenotype
If we’re going to talk about breeding, then we need to discuss genetics, a widely misunderstood subject. So let’s start off by defining some terms [2].
Population, or more specifically, the “breeding population”: a population of organisms within which free interbreeding takes place and evolutionary change may appear and be preserved. Mother Nature does not recognize “species;” species names are merely a human construct invented by those who wish to organize everything into categories. In nature, there only exist “populations” of interbreeding individuals sharing a common gene pool, and whether or not they are species, subspecies, or races is a matter of debate for the taxonomists.
The phenotype of an individual honey bee, a colony, or a population is the set of observable characteristics (size, color, honey production, wintering ability, defensiveness).
The genotype (the “genetics”) of a bee or colony is the set of inherited genetic instructions encoded in its DNA. But not all organisms with the same genotype look or act the same, because appearance and behavior are modified by environmental and developmental conditions. Likewise, not all organisms that look alike necessarily have the same genotype.
Genes: when we refer to two bees having different “genes,” what we really mean is that they have two different forms (variants) of the same gene. Different forms of a gene are called alleles.
Both in natural selection and in traditional selective breeding, selection is applied to the expression of the phenotype, rather than the genotype [3], since it is the phenotype that directly interacts with the environment. Absent genetic techniques, all that we can describe as differences between any two bees, colonies, or races of bees are phenotypical variations. In nature, there is often a continuum (rather than discrete steps) of phenotypes, as well as of genotypes, in the individuals composing a species over its range [4](Fig. 2).
Figure 2. Note the continuum of abdominal coloration patterns in the bees of this hive, ranging from black/gray to nearly all golden, with various widths and darkness of the banding. I’m happy to see such a mix of colorations (phenotype) in my hives, as a visible proxy for genetic diversity (genotype).
Taxonomists describe nearly 30 races (subspecies) of honey bee over the species’ natural range from the tip of Africa, up through the Middle East and Mediterranean, and northward into Europe. Traditionally, these races were classified by physical characteristics, such as color, size, behavior, and morphometric analysis (measuring wing venation and body part proportions). More recently, we can use genetic tools to differentiate these races by comparing the variety of forms (alleles) of various selected genes [5]. In the various races of bees, certain alleles are said to be “fixed” (or “nearly fixed’); that is, only one form of that particular allele is found in that particular breeding population. (Note the lack of fixation in the coloration patterns of the bees in the photo above. In the identification of wasps (in which the coloration patterns are fixed), such differences would be found only in separate species [6].
When we speak of a population having genetic diversity, what we mean is that that population carries a diverse mixture of non fixed alleles. This diversity is critical for species survival in the wild, since in nature, things change. There are droughts, heat waves, hordes of locusts eating all the forage, unusually frigid winters, late springs, pathogen epidemics, or other conditions that favor colonies of bees possessing certain combinations of alleles over others. It is only through this genetic and phenotypic diversity that a portion of a species’ population is able to survive such extreme or unusual events, or adapt to other changes in the environment. This concept also applies to the individual bee colony:
Practical note: the success of a colony is dependent not only upon the genotype and phenotype of its queen (such as her ability to lay a great many eggs and her production of adequate pheromones), but even more so upon the combination of phenotypes of the various patrilines of workers (the collective offspring from each of the individual drones with which the queen mated). Colonies without a diversity of patrilines exhibit poor disease resistance and winter survival [7].
Now I’ve got one more term to throw at you:
Epigenetics: the heritable changes in gene expression that are not caused by changes in the DNA sequence; the term can also be used for an organism’s regulatory responses to the environment that may be heritable for one or more generations.
So what does that mean? Every bee comes with a genetic manual encoded in its DNA. That manual contains “instructions” with numerous options as to how to build every part of its body, how to make every physiological system work, and for every aspect of its behavior. Its epigenetics tell it which options to apply, based either upon the bee’s heritage, feeding as a larva, its exposure to pheromones, or to other environmental cues. The end result is that the bee’s phenotype (physical form) is a result of the epigenetic expression of its genotype. Let me use the epigenetic differentiation of the worker, queen, and drone bees as an example (Fig. 3):
Figure 3. The obvious phenotypic differences between workers, queens, and drones are not due to genetics, but rather due to epigenetic regulation of the development of the immature bee. Illustration from [[i]].
[i] Illustration from Kauffeld, NM (1980) Seasonal cycle of activities in honey bee colonies. In Beekeeping in the United States, Agriculture Handbook Number 335.
The worker, queen, and drone above are clearly different organisms in size, physiology, morphology (shape and structure), longevity, reproduction, and behavior (only workers forage or sting in colony defense). The amazing thing is that a fertilized egg has to genetic potential to become any of the three. The difference in their developmental paths from egg to sexual adult is not due to genetics, but rather to epigenetics.
Unlike humans, in which maleness is determined by the inheritance of a sex-determining Y chromosome, in bees (which have no sex chromosome) the default development of any fertilized egg is to become a male (yes, fertilized eggs can develop into viable diploid drones [9]). The egg will become a female only if it inherited two different variants of the “sex allele” at one specific gene locus on chromosome 8 (the complementary sex determiner (csd) gene). Haploid (unfertilized) eggs would of course have only have one sex allele, since they’d have only a single set of chromosomes, and thus always become males. Surprisingly, the csd gene is not even directly involved in the feminization process—the presence of two different alleles at this “sex locus” is merely a trigger for the cell to epigenetically activate another gene (called feminize) [10], which then starts the process of feminization.
My point is that since the csd gene doesn’t code for any proteins involved in actually growing the bee, that means that for all intents and purposes a worker, queen, or drone are genetically identical, and it is only the epigenetic regulation that makes them develop differently. In the female castes, this regulation is based upon what the larvae are fed by the nurses (who choose whether a larva will become a worker or a queen) [11].
In other words, the bee is akin to the stem cells in your body—it can develop into any number of forms. Such an organism is said to exhibit phenotypic plasticity. And the honey bee exhibits such plasticity at both the individual and at the colony level. As observed by Weiner and Toth [12]:
Phenotypic plasticity is an important biological phenomenon that allows organisms with the same genotype to respond adaptively to variable biotic and abiotic environments.
In some of my previous articles on pheromones and the division of labor in the colony [13], I showed how the colony responds to environmental cues such as nectar flows, pollen dearth, status of the queen, pheromones, and illness. Such day-to-day or region-to-region adaptability is largely brought about by the epigenetic up- or down-regulation of the same set of genetic instructions. Of considerable interest is Robert Paxton’s experimentation (described in a fascinating video [14]) in which he found that one species of sweat bee can live either as a solitary bee or as a eusocial colonial bee, dependent upon the temperature of the environment!
Practical application: simple differences in regulatory triggers (e.g., influence of temperature [15], pheromones, components in the jelly fed to larvae, or the activation of a single regulatory gene) can result in major differences in the shape, structure, and behavior of genetically identical bees. My point is that it may not take much genetic evolution in order to result in big differences in the phenotype of the honey bee. To the early beekeepers selecting for better bees, they needn’t have changed the genome to any great extent—they only needed to select for some minor epigenetic regulation of the existing genome. I will return to this important point next month.
A very recent study is of interest. Harpur, et al [16] undertook a comprehensive population genomic study of the honey bee by sequencing the genomes of 40 individual bees from different geographic regions. They found that the genes coding for proteins expressed solely by workers have evolved at a greater rate than those for queens, especially those associated with division of labor, the nursing of brood, worker behavior, worker sensory responses (such as to pheromones or other environmental cues), cognition, nervous system development, metabolism, and steroid hormones.
Practical application: so it is likely not important to select for queen characteristics (size, shape, color), but rather to select for the performance of the entire breeding population of colonies from which to pick your breeders (in my own breeding program, I don’t even bother to see what the queen looks like).
Look, I’m at about my limit of understanding of all this, but every single aspect of bee reproduction suggests to me that it is set up for three things:
- Maintaining genetic diversity. The ecological success and adaptability of the honey bee is all about its genetic diversity, not only at the population level, but also at the colony level.
- Rapid adaptability. The high genetic recombination rate of the species [17] coupled with the haploidly of the drones (which effectively weeds out maladaptive allelic combinations [18]), the multiple matings by the queen, the amazing epigenetic adaptive plasticity of the species, and the rapid rate of subspeciation all cry out adaptability.
- Recovery from decimation events. Catastrophic events such as drought, forest fire, extreme winters, unfavorable seasons, or epidemics of infectious disease can decimate a regional population of bees. But due to the genetic reservoir of any surviving colonies (stored in the diverse semen in the queens’ spermathecas), the swarms issued from the survivors have the potential to reestablish much of the original genetic (and epigenetic) diversity of the former population—each colony of bees is essentially a “genetic ark.” Estoup [[19]] observes that “the average heterozygosity [genetic diversity] of a population [of honey bees] can be estimated from a single colony with fairly good precision.”
Practical application: when we practice selective breeding of the honey bee, we are fighting the nature of the beast. The extraordinary ecological success and adaptability of the honey bee is all about maintaining genetic diversity; selective breeding, on the other hand, results in loss of such diversity. Perhaps we should seriously question whether we have been helping or hurting the honey bee as a species. Again, I will return to this subject later.
The Gene Ocean
Apis mellifera doesn’t just have a gene pool—it has (or had) a “gene ocean” to draw from. Those many races of bees stretching from South Africa to the Baltic form a continuum of shared genetics [20]. Hepburn [21] explains:
Populations of honeybees previously thought to be homogenous and thus defined as races or subspecies, actually emerge as a potpourri of independently oscillating traits within a continuous metapopulation. In this respect, honeybee populations confirm the tendency noted for other animals and plants in which genetically independent characters show independent geographical variation and have the capacity to recur in more than one geographical area.
Just as the waters of the ocean swirl and slowly mix, there is a flow of alleles (commonly called “gene flow”) throughout the vast population of Apis mellifera. And as new mutations, viral integration of genes [22], novel genetic recombinations, and heritable epigenetic factors occur, these may meld into that flow and add to the overall genetic diversity of the species as a whole. Within any race of honey bees, there are subpopulations that noticeably differ; and in the areas between any two races of bee, there are zones of hybridization in which novel genetic combinations are continually tested for fitness.
OK, you ask, what’s this got to do with the domestication of the honey bee? I’m getting to that. The process of domestication directly affects the…
Size and Genetic Diversity Of The Breeding Population
Now we get to the crux of the issue:
Practical application: the ability of a population of bees to adapt to changes in the environment or to novel pathogens is largely dependent upon its genetic diversity (see sidebar [citations [23]).
In interest of simplicity, I’m going to use the term “genetic diversity” to refer to all heritable factors, genetic and epigenetic.
At this point in time, there is a paradigm shift going on in biology as we begin to understand that the interpretation of genome into phenotype is done by epigenetic regulation [a].
The genetic code is like having the mind-boggling 74,000-page U.S. tax code sitting on your lap. Epigenetics is like having a business consultant, accountant, attorney, and tax advisor there to tell you how to apply that code to your advantage.
Epigenetic factors may be temporary (the responses to pheromones [b], for the life of the organism (effects of pesticide exposure or larval temperature or nutrition), or transgenerational (for one or more generations; or “fixed” for all generations)[c].
And there are sometimes huge differences in transgenerational epigenetic inheritance dependent upon the sex of the contributing parent [d]. We are only beginning to understand the complexity of epigenetics, and I am no expert; but they certainly play a large role in honey bee biology and breeding!
The maintenance of the genetic diversity in a population is dependent upon several factors, notably:
- The rate of mutation and recombination (the swapping of genes in the process of germ cell formation),
- gene flow within the breeding population (and from without, including hybridization), and
- the total ”Effective Population Size” (Ne).
I’ve already mentioned that bees have a high rate of mutation and gene flow. But what do I mean by Effective Population Size?
When the gene pool of a population of interbreeding organisms is reduced in size (even for a single generation), either due to decimating catastrophic events, geographical isolation (as by climate change isolating populations between oceans, mountains, deserts, or glaciers), or the invasion of a breeding population into new territory, the genetic diversity of that gene pool is often reduced to a “gene puddle” (not a scientific term). Such an event is called a “genetic bottleneck” [24], which reduces the “effective population size.”
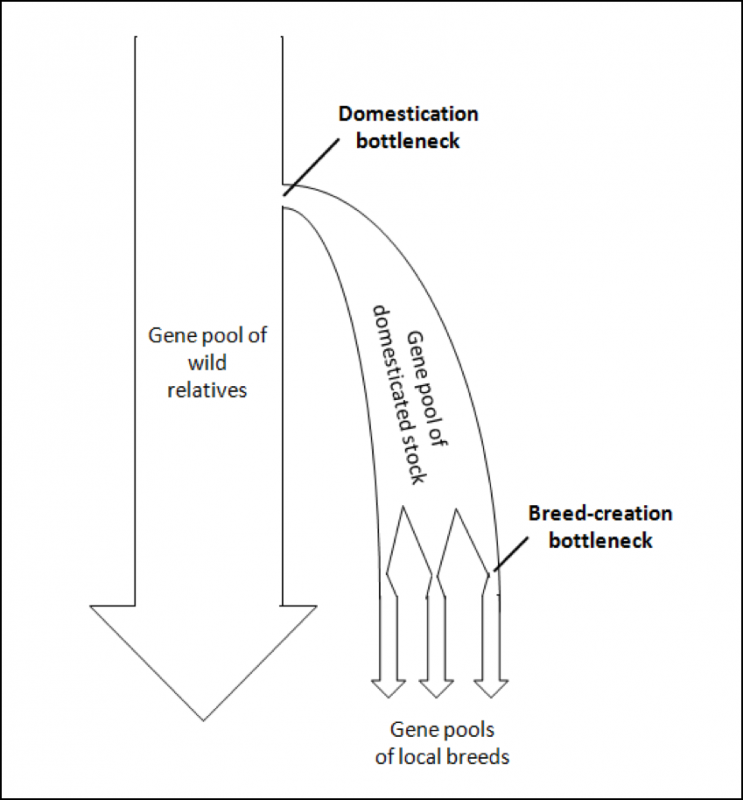
To illustrate these bottlenecking processes, I’ve modified a visual illustration from a paper by Wang [25] (Fig. 4).
Figure 4. A generic model of the genetic bottlenecks resulting from the selective breeding involved in the process of domestication. At each bottleneck, the effective population size is reduced. The question then is, to what degree have our current breeds of bees lost genetic diversity? Graphic modified from [[i]].
[i] Wang (2014), Ibid.
The Consequences Of Bottlenecks
Following any bottlenecking event, the population must rebuild. The problem is that such rebuilding is limited by the genetic diversity of the founding population (which was only a portion of that of the original population). Even worse, as the population rebuilds from the founders (either due to natural causes or human selection), at each generation, some alleles, due to either randomness of matings or human selection, tend to be lost. This phenomenon is termed genetic drift, the result being that populations established from small founder populations tend to suffer from loss of genetic diversity—the smaller the original (and the maintained) population, the greater the loss of diversity. One resulting negative effect is called inbreeding depression, due to the expression of unmasked deleterious recessive alleles (a subject to which I will return).
Such reduction in the effective population size is especially problematic with species that have haploid males with a sex determining gene (case in point, the honey bee) [27]. Colonies headed by an inbred queen suffer from low brood viability [28], due to the development of “diploid drones” when a fertilized egg did not inherit two different sex alleles from its parents. Since a queen is limited in the number of eggs that she can lay a day, reduced viability of the resulting brood can greatly reduce the ability of a colony to grow and store honey.
Practical application: this is why you want to see solid brood patterns in a hive.
But that’s not the half of it! A far more important deleterious effect of inbreeding is that the resulting population has a greatly reduced ability to fight infectious diseases (for an overview of this phenomenon, I suggest [29]). The beneficial effect of genetic diversity to slow the spread of infectious diseases is commonly used in agriculture [30]. In a worst-case situation, in which all organisms in a population are genetically identical (as in typical monocultures of cloned cultivars) the first pathogen able to infect one individual can quickly spread to all others in epidemic fashion (think of the Irish potato famine, the Gros Michel banana, or the decimation of the Innuit by the Spanish flu). On the other hand, if a population of organisms is genetically diverse, slight differences in proteins and immune responses among individuals inhibits the transmission rate of pathogens, limiting the progression of epidemics.
Practical application: of major and direct importance to beekeepers, especially in these days of globalization and the transport of pathogens and parasites from one bee population to another, is that bee populations with a limited genetic toolbox exhibit less vigor, decreased ability to fight epidemics of parasites or infectious diseases [31], and less ability to evolve and adapt.
Cases in point:
- The vigorous untreated Southern California colonies that so impressed me.
- The rapid natural evolution of varroa resistance in both unmanaged races of bees in South Africa (taking place in 4- 6 years, as compared to not yet happening after decades of varroa presence in Europe) [32].
So How Much Genetic Diversity Have We Lost?
Good question! This is a theme that has been raised for a number of years by Steve Sheppard, Sue Cobey, and Debby Delaney (I’ll return to their findings later).
One possible indicator of genetic diversity is to look at the number of sex alleles in the population (since these are all variants of a single gene). I’m not sure if this is the best indicator, since this gene appears to mutate at a relatively high frequency (on an evolutionary time scale). Given that caveat, the diversity (or lack thereof) of sex alleles in our domestic bee populations may be an indicator of the degree to which our selective breeding has narrowed down the gene pool.
So how much genetic diversity was lost as Apis mellifera migrated into Europe to found the current races of bees, and then further lost as beekeepers selected specific manageable stocks?
Early estimates [33] of the number of sex alleles present in the various races of bees ranged from 8–12. Later, Adams [34], sampling 90 Brazilian colonies consisting of freely-mated hybrids between A. mellifera adansonii and A. mellifera ligustica, with at least 500 other hives within flight range, came up with an estimate of about 20 sex alleles for this larger hybrid population.
The above estimates suggest that the presence of 8–20 sex alleles may be the norm for managed populations of European bees. So how does that compare to the diversity in the wild bee population as a whole? Lechner [35] took on this task, stating that:
When studying the population dynamics of csd alleles in honey bees, one is faced with the history of enormous anthropogenic influence (bee management) in many parts of the world. As detailed herein, we circumvented this problem by sampling csd sequences from localities in Kenya (East Africa), where the anthropogenic influence is negligibly low.
She sampled 2 workers each from 10 hives, from three regions in Kenya, as well as hives from 3 U.S states, Brazil, Israel, and Australia. She found that the calculated number of sex alleles was 53 on a local basis, and likely about 90 worldwide, suggesting long-term effective population sizes ranging from 12,000 to 34,000 colonies.
The above findings are worth thinking about. If the norm for wild bees in their homeland is to have a diversity of 50 sex alleles (with a possible 90 in the entire Apis mellifera population), and if we (prior to varroa) were finding only about 10 alleles, what other genetic diversity have we lost in the process of domestication?
Practical application: Let me jump ahead about 3000 years for a moment, to our current stocks of bees in the U.S., founded from the limited original importations of a few races of bees (bottleneck 1), then selected over the years by breeders (bottlenecks 2), then having the effective population size again greatly reduced when tracheal and varroa mites wiped out most of our domestic and feral stocks (serious bottlenecks 3). How have those three bottleneck events affected the vigor of the bee populations that we keep nowadays?
Final Notes
I’ll end this article with the above open question about the loss of genetic diversity in our bee stocks. But I’ll continue next month with another question: Is there really a difference between domesticated and wild honey bees?
Again, I’m doing my best here at attempting to summarize our state of knowledge of this extremely interesting but complex subject—please let me know if I’ve made any errors in my interpretation of the science (some of the genetics papers read like Greek to me)!
I also wish to reiterate that I am not criticizing our queen breeders or domesticated stocks in any way (if I wanted milk, I sure wouldn’t want to try to get it from wild goats). But I think that it would be wise for us to fully grasp any inherent negative consequences of the selective breeding of the honey bee, especially with regard to its adaptation to the varroa mite. I will of course continue next month.
Acknowledgements
Thanks as always to Peter Borst in research assistance. And thanks to those who donate to ScientificBeekeeping.com to allow me to continue my research and writing.
Footnotes And Citations
[1]. Chaudhary, B (2013) Plant domestication and resistance to herbivory. International Journal of Plant Genomics http://www.hindawi.com/journals/ijpg/2013/572784/
[2] I’ve used Wikipedia and Merriam-Webster as sources for these definitions.
[3] Recent advances in technology now allow us to use Marker Assisted Selection and/or genetic engineering to directly select for specific alleles (forms of a gene) or even to swap genes between species.
[4] This is called continuous, as opposed to discontinuous, variation over a range.
[5] Researchers often look at SNPs (single nucleotide polymorphisms), which are substitutions in the DNA code that do not necessarily affect anything, but are useful identifiers that can track inheritance and evolution.
[6] http://www.discoverlife.org/mp/20q?guide=Vespinae
[7] Seeley, TD and DR Tarpy (2007) Queen promiscuity lowers disease within honeybee colonies. Proc. R. Soc. B. 274: 67-72. Open access.
Matilla, HR and TD Seeley (2007) Genetic diversity in honey bee colonies enhances productivity and fitness. Science 317: 362-364. Open access.
[8] Illustration from Kauffeld, NM (1980) Seasonal cycle of activities in honey bee colonies. In Beekeeping in the United States, Agriculture Handbook Number 335.
[9] Polaczek, B, et al (2000)A new, simple method for rearing diploid drones in the honeybee (Apis mellifera L.). Apidologie 31: 525–530. Small colonies will naturally rear to adulthood diploid drones in fall.
Herrmann M1, Trenzcek T, Fahrenhorst H, Engels W. (2005) Characters that differ between diploid and haploid honey bee (Apis mellifera) drones. Genet Mol Res. 2005 Dec 30;4(4):624-41.
[10] Gempe, T and M Beye (2009) Sex determination in honeybees. Nature Education 2(2):1 http://www.nature.com/scitable/topicpage/sex-determination-in-honeybees-2591764#
[11] Kucharski R, et al (2008) Nutritional control of reproductive status in honeybees via DNA methylation. Science 319: 1827–1830. http://www.sciencemag.org/content/319/5871/1827.full
Kamakura, M (2011) Royalactin induces queen differentiation in honeybees. Nature 473(7348):478–483). Open access.
Guo X, et al. (2013) Recipe for a busy bee: MicroRNAs in honey bee caste determination. PLoS ONE 8(12): e81661. Open access.
[12] Weiner, SA and AL Toth (2012) Epigenetics in social insects: a new direction for understanding the evolution of castes. Genetics Research International http://www.hindawi.com/journals/gri/2012/609810/
[13] See the An Adaptable Workforce, The Primer Pheromones, and Sick Bees 2.
[14] Paxton, R (2013) Origins and evolutionary history of the honey bee. http://www.youtube.com/watch?v=Wtm8URk-V9A&feature=youtu.be A fascinating video presentation on bee evolution.
[15] Becher, MA, et al (2009) Pupal developmental temperature and behavioral specialization of honeybee workers (Apis mellifera L.). Journal of Comparative Physiology A 195(7): 673-679.
[16] Harpur, BA, et al (2014) Population genomics of the honey bee reveals strong signatures of positive selection on worker traits. PNAS 111(7): 2614–2619. http://www.pnas.org/content/111/7/2614.full?tab=author-info
See also: Simola, DF, et al (2013) Social insect genomes exhibit dramatic evolution in gene composition and regulation while preserving regulatory features linked to sociality. Genome Res. 23: 1235-1247. Open access.
[17] Beye, M, et al (1999) unusually high recombination rate detected in the sex locus region of the honey bee (Apis mellifera). Genetics 153: 1701–1708.
[18] “Should erosion of genetic diversity occur, haplodiploid organisms might be expected to suffer less from inbreeding depression than diploids because deleterious recessive alleles may be purged through selection on the haploid sex.” Ellis, JS (2006) Extremely low effective population sizes, structuring and reduced genetic diversity in a threatened bumblebee species, Bombus sylvarum (Hymenoptera: Apidae). Molecular Ecology 15:4375–4386.
[19] Estoup, A, et al (1995) microsatellite variation in honey bee (Apis mellifera L.) populations: Hierarchical genetic structure and test of the infinite allele and stepwise mutation models. Genetics 140 679-695. Open acess.
[20] Diniz-Filho, JAF, et al (2000) Spatial analysis of morphological variation in African honeybees (Apis mellifera L.) on a continental scale. Apidologie 31: 191–204. Open acess.
Radloff, SE.; Hepburn, HR.; Fuchs, S., (1998) Ecological and morphological differentiation of the honeybees, Apis mellifera Linnaeus Hymenoptera Apidae, of West Africa. African Entomology. March; 61: 17-23.
[21] Hepburn, HR and SE Radloff (1998) Honeybees of Africa. Springer-Verlag.
[22] Maori, E, et al (2007) Reciprocal sequence exchange between non-retro viruses and hosts leading to the appearance of new host phenotypes. Virology 362: 342–349. Open access.
[23] a. Jablonka E and MJ Lamb (1998) Epigenetic inheritance in evolution. Journal of Evolutionary Biology, 11, 159-183. http://onlinelibrary.wiley.com/doi/10.1046/j.1420-9101.1998.11020159.x/pdf
- Alaux, C, et al (2009) Regulation of brain gene expression in honey bees by brood pheromone. Genes, Brain and Behavior 8: 309–319. Open access.
Alaux, C, et al (2009) Honey bee aggression supports a link between gene regulation and behavioral evolution. PNAS 106: 36. http://www.pnas.org/content/106/36/15400.full
- Lucia Daxinger, Land E Whitelaw (2010) Transgenerational epigenetic inheritance: More questions than answers. Genome Res. 20:1623-1628. http://genome.cshlp.org/content/20/12/1623.full
- Kappeler, L and J Meaney (2010) Epigenetics and parental effects. BioEssays 32(9): 818–827.
[24] http://evolution.berkeley.edu/evosite/evo101/IIID3Bottlenecks.shtml
[25] Wang, G-D, et al (2014) Domestication genomics: evidence from animals. Annu. Rev. Anim. Biosci. 2: 65–84.
[26] Wang (2014), Ibid.
[27] Zayed, A (2004) Effective population size in hymenoptera with complementary sex determination. Heredity 93: 627–630. Open access.
[28] Mackensen, O (1951) Viability and sex determination in the honey bee (Apis mellifera L.). Genetics3 6: 500-509. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1209536/pdf/500.pdf
[29] O’Brien, SJ and JF Evermann (1988) Interactive influence of infectious disease and genetic diversity in natural populations. Trends in Ecology & Evolution 3(10): 254-259. A fascinating paper on the interactions between host genetic diversity, bottlenecking events, and viral epidemics. (Broken Link!) http://dobzhanskycenter.bio.spbu.ru/pdf/sjop/MS155_.pdf
[30] Zhu Y, et al (2000) Genetic diversity and disease control in rice. Nature 406, 718-722. Open access.
Garrett, KA & CC Mundt (1999) Epidemiology in mixed host populations. Phytopathology 89, 984-990. Open access
[31] Reviewed in Wilson-Rich, N, et al. (2012) Within- and across-colony effects of hyperpolyandry on immune function and body condition in honey bees (Apis mellifera). Journal of Insect Physiology 58: 402–407.
[32] Allsopp, M (2006) Analysis of Varroa destructor infestation of Southern African honeybee populations. M.S. Thesis, Univ. of Pretoria. http://upetd.up.ac.za/thesis/available/etd-08082007-153050/unrestricted/dissertation.pdf A must read for anyone interested in breeding bees for varroa resistance.
[33] Reviewed by: Adams, J, et al (1977) Estimation of the number of sex alleles and queen matings from diploid male frequencies in a population of Apis mellifera. Genetics 86: 583-596. http://www.genetics.org/content/86/3/583.full.pdf
[34] Ibid.
[35] Lechner, S, et al (2014) Nucleotide variability at its limit? Insights into the number and evolutionary dynamics of the sex-determining specificities of the honey bee Apis mellifera. Mol Biol Evol. 31(2): 272–287. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3907057/?report=classic