What’s Happening To The Bees? – Part 5: Is There A Difference Between Domesticated And Feral Bees?
CONTENTS
But Aren’t All Feral Bees Nothing More Than Escaped Swarms?
Other Domesticated Animal Models
The Mechanics Of Domestication
The Establishment Of A Feral Population
Gene Flow Between Managed And Wild Stocks
What’s Happening To The Bees? – Part 5:
Is There A Difference Between Domesticated And Feral Bees?
Randy Oliver
ScientificBeekeeping.com
First published in ABJ in June 2014
I mentioned previously how impressed I am by the difference in vigor between the Southern California feral bees and commercial domesticated stock. I also made the bold statement that the honey bees produced by most commercial queen breeders could be considered to be domesticated animals. Such statements call for some serious supporting evidence. Is there truly a difference between domesticated and wild/feral honey bees? The obvious question is:
But Aren’t All Feral Bees Nothing More Than Escaped Swarms?
In areas in which the honey bee was not endemic (native), this would initially of course be the case. But once those introduced swarms set up housekeeping in hollow trees, we then have two key questions to answer:
Question #1: Can those introduced feral populations remain self sustaining, or does their continued survival depend upon a recurrent influx of escaped swarms from managed apiaries? And if those feral populations are indeed self sustaining,
Question #2: Do established feral populations differ genetically from the sympatric (living in the same area) population(s) of managed bees? In other words, do the genetics of the feral populations simply reflect that of the swarms and drones to which they are continually exposed, or do the ferals maintain genetic integrity independent of the managed bee populations?
Practical application: answering the latter question is critical if we are to evaluate whether there is any substance to the argument that we beekeepers should view the feral populations of honey bees as potential sources of genetically useful “survivor stock.”
Other Domesticated Animal Models
Honey bees are hardly the only animal (or plant) that has been domesticated (or perhaps I should use the term “managed”) by humans [1], so as a first step in answering the above questions, perhaps we should see what’s been found regarding other common domesticated species. Charles Darwin wrote two fascinating volumes about his observations on the phenotypical [2] variation in species under domestication [3], from which I’ll share some excerpts:
From a remote period, in all parts of the world, man has subjected many animals and plants to domestication or culture…Although man does not cause variability and cannot prevent it, he can select, preserve, and accumulate variations given to him by the hand of nature in almost any way which he chooses; and thus he can certainly produce a great result. Selection may be followed either methodically or intentionally, or unconsciously and unintentionally. Man may select and preserve each successive variation, with the distinct intention of improving and altering a breed in accordance with a preconceived idea; and by thus adding up variations, often so slight as to be imperceptible by an uneducated eye, he has effected wonderful changes and improvements. It can, also, be clearly shown that man, without any intention or thought of improving the breed, by preserving in each successive generation the individuals which he prizes most, and by destroying the worthless individuals, slowly, though surely, induces great changes.
Jumping ahead a hundred years, a more modern scientist [4], using today’s more technical terminology, observes:
In contrast to wild species, which have typically evolved phenotypes over long periods of natural selection, domesticates rapidly gained human-preferred agronomic traits in a relatively short-time frame via artificial selection.
Ah, the test of time. Any species extant today is by definition a “survivor”—the product of an unbroken line of survivors dating back to the beginning of life (any single break and that line would have gone extinct). On the other hand, any and all domesticated breeds of animals have existed for only a few thousand years, and only by the grace of man’s facilitation of their niche.
Riding our time machine back to Darwin, he keenly observed that those “wonderful changes and improvements” come at a cost—the ability to survive in the wild.
As the will of man thus comes into play, we can understand how it is that domesticated breeds show adaptation to his wants and pleasures. We can further understand how it is that domestic races of animals…often exhibit an abnormal character, as compared with natural species; for they have been modified not for their own benefit, but for that of man.
So key to my initial questions, is what happens when managed honey bees (or other livestock) escape captivity and attempt to revert to life in the wild? Of interest are Darwin’s lengthy observations on such “reversion” of domesticated species back to wild type characteristics:
Reversion To Wild
A feral animal (from Latin fera, “a wild beast”) is an animal living in the wild but descended from domesticated individuals [5]. Darwin points out that:
…we have seen that characters often reappear in purely-bred races without our being able to assign any proximate cause; but when they become feral this is either indirectly or directly induced by the change in their conditions of life.
Even purebred breeds hide aspects of their genetic heritage from us [6]. Two factors may cause these hidden traits to come to the fore: environmental factors or crossbreeding.
Darwin noted that the first generation of escaped domestic animals might exhibit minor phenotypical changes, in response to environmental cues, such growing more hair when it got cold. But what really caught his (and should catch our) attention was what could happen when two purebred breeds were “crossed.” The resulting hybrid [7] offspring are often noticeably different from either parent:
With crossed breeds, the act of crossing in itself certainly leads to the recovery of long-lost characters, as well as those derived from either parent form…From what we see of the power and scope of reversion, both in pure races, and when varieties…are crossed, we may infer that characters of almost every kind are capable of reappearing after having been lost for a great length of time.
He further noted that the most common form of reversion, “almost universal with the offspring from a cross, [is to go back] to the characters proper to either pure parent form.”
I share Darwin’s fascination with this clearly observable phenomenon.
Practical application: when queens of a managed stock mate with drones of other stocks, the resulting offspring may exhibit wild-type traits that had long lain dormant. Could this be the basis of a wild-type feral population, originally founded by escaped swarms from a variety of domesticated lines?
The Mechanics Of Domestication
Clearly, species have the ability to carry “hidden” genes for many generations. How can this be?
The process of selective breeding creates genetic bottlenecks, in which some allelic diversity is lost, and the frequency of some deleterious alleles is increased. But for the most part, any domesticated breed retains a full complement of the wild-type genes necessary for basic bodily functions and behaviors [8]. The difference is that in domesticated animals, the expression of certain genes is differentially regulated.
Nature is very conservative with the genetic code. Honey bees share a set of generic genes common to all insects for most of their operational systems (and indeed share some with humans). The genetic differences between species are often trivial (for example, the DNA of any reader of this article is 98.8 percent identical to that of a chimpanzee (and not just because we’re beekeepers) [9]). What makes species and races different is the regulation of those genes.
As a crude analogy, think of a wild-type animal as being a tough military-grade 4WD pickup truck. Different domesticated breeds of that animal are analogous to consumer options on the basic truck model, such as a different body type, paint color, larger tires, leather upholstery, radio and air conditioning, or a supercharged engine. All those models are simply minor variations of the basic truck to suit the customer—none involve fundamental changes in the chassis or drive train.
Given an assortment of models to strip and reassemble, a team of backyard mechanics could easily recreate the wild-type truck (or perhaps an improvement). And this is what appears to happen when an assortment of breeds of a domesticated animal escape into the wild—their offspring tend to revert to wild type. The fundamental genes were always there; the crossing of strains simply changed the expression of those genes back to “normal.”
Practical application: the reversion to “wild type” does not appear to require major genetic reengineering—rather, all that may be necessary is the tweaking of only a few critical regulatory genes.
My impression is that this is exactly what happens when one crosses races of Apis mellifera or when feral colonies start swapping drones. The result is the production of a more ancestral wild type bee population, with a larger degree of genetic variation. This population now experiences selective pressure from nature, rather than from man, which strongly selects for genetic and epigenetic combinations that are successful in that particular environment (fitness). Hence, we may find feral populations, despite the fact that they were founded from stocks selected for domestication, reverting to wild-type characteristics (Fig. 1).
 Figure 1. An escaped swarm of domesticated stock can easily take up residence in a hollow tree and fill the cavity with combs. But can a bee bred for almond pollination and the filling of quadruple-high Langstroth hives, facilitated by feeding and medication by its keeper, survive for long in natural tree cavities in the wild?
Figure 1. An escaped swarm of domesticated stock can easily take up residence in a hollow tree and fill the cavity with combs. But can a bee bred for almond pollination and the filling of quadruple-high Langstroth hives, facilitated by feeding and medication by its keeper, survive for long in natural tree cavities in the wild?
A point of interest is that when one crosses a gentle strain of the Dark Bee (A. m. mellifera) with gentle Italian (A. m. ligustica) stock, the hybrid offspring are often reported to be fiercely “hot.” This suggests to me that defensiveness is an innate trait that has been epigenetically downregulated in domestic stocks; hybridization may allow the default trait to express itself fully (and painfully).
It certainly appears that feral populations of bees founded from escaped swarms can indeed be self sustaining. Strong evidence in support of this comes from a study by a team headed by Australian researcher Ben Oldroyd [10]. They found that established feral colonies reproduced (sent out swarms) at a higher rate than their death rate, meaning that their populations would be mathematically self sustaining.
The above study followed necessary scientific rigor. But pretty much every beekeeper is aware of the fact that Apis mellifera has rapidly invaded and colonized virtually any continent or island to which they’ve been introduced, including varroa-free Australia, which continues to support a robust feral population (of “black bees”) independent of managed stocks (of “yellow bees”) [11].
Practical application: I find no support whatsoever for the argument that feral populations are dependent upon continual support from escaped swarms of managed bees (in fact, as I’ll argue later in this series, the nearby presence of managed bees actually may harm the feral or wild populations of honey bees).
The Establishment Of A Feral Population
Just because the ferals were founded from inbred domesticated bees does not mean that they can’t quickly revert back to a more genetically diverse wild type population. But it’s hardly a straightforward process, as there are a number of sometimes competing factors involved:
- A small founder population invariably carries only a portion of the totality of the original species’ gene pool. This bottlenecking event may or may not reduce the invasive success of the introduced species in the new habitat [12] (many intentional introductions of various species fail). In the case of honey bees, of the twenty-some races of Apis mellifera, only eight were originally introduced into the U.S. [13]. Thus, our feral population started with only a fraction of the “gene ocean” present in the bees’ homeland.
- Due to the haploid/diploid mating system and the sterility of diploid drones, nature strongly selects against inbred lines of bees. This can negatively affect the establishment of a founder population of honey bees. On the other hand, it can also quickly weed out nonadaptive (less fit) genetic combinations.
- Bees have an unusual potential for skirting the above “founder effect,” since each queen carries not only the genes (and epigenes) of her mother, but also those of all the drones that she mated with. So even a single colony may carry a wide diversity of alleles to start with. The problem is in getting this diversity into the next generation, since it can occur only via daughter queens, not directly from that stored sperm (since drones come from unfertilized eggs).
- Founder populations that rapidly expand in an environment may not suffer from loss of allelic diversity [14]. But again, bees are a special case. Keep in mind that the mother queen leaves with the first swarm, leaving behind a colony headed by one of her daughters. But the genetic diversity of the drone pool available to that first generation of daughters would be limited to that of the foundress mothers alone, and couldn’t take advantage of the diversity of the sperms in their spermatheca [15]. But each virgin daughter produced by a foundress queen would likely have been sired by sperm from a different father, so the most diversity would be preserved if the foundress queens, in their remaining lifetimes, were able to produce multiple swarms and afterswarms.
- And then such an expanding founder population may suffer from random genetic drift—permanently losing some beneficial alleles, or increasing the proportion of some deleterious alleles [16].
- Additional introduction or immigration of stock can prevent that loss of diversity from genetic drift [17]. In the U.S., numerous early imports, coupled with the relatively recent importation of Buckfast, Yugo, Australian, and Primorsky Russian bees, legal importations of semen by Cobey and Sheppard, other illegal importations by beekeepers, and the invasion of Africanized stock have all added additional alleles to the population.
- Finally, the introduction of multiple races of a species may actually result in greater genetic diversity in the resulting feral population than that found in the native populations in their homelands [18]. And this appears to be the case with honey bees. A recent study by Harpur [19] “found that managed honey bees actually have higher levels of genetic diversity compared with their progenitors in East and West Europe, providing an unusual example whereby human management increases genetic diversity by promoting admixture.”
Practical application: Harpur’s findings in populations of managed bees in Ontario and France are surprising. Who would have thought that despite the bottlenecking due to the small founding populations, the genetic drift due to strong selection by bee breeders, and the fact that a relative handful of queen breeders produce the majority of queens from only a few hundred mothers each year [20], that managed bees would exhibit greater genetic diversity than their wild counterparts in Africa, West-, and Eastern Europe? The question now remains, how does the genetic diversity of the feral population in North America compare to that of our managed bees? We’ll get to that soon…
Quid Pro Quo
One wouldn’t think of honey bees as being fully domesticated, since escaped swarms easily survive in the wild (as feral bees). Or do they?
Domestication of a species typically creates (at the genetic level) a dependency upon human husbandry such that the animal finds it difficult to live in the wild [21]. The reason for this is that when we select for certain characteristics desirable by humans, there is a quid pro quo—something must be given up for something in return (a zero sum game). That something given in return for high honey production, color, or docility may be a reduced ability to survive in the wild.
For example, humans have bred the silkworm for thousands of years, developing hundreds of varieties selected for higher silk production (more than tripling that production since the year 1800 [22]). But such improvement came at a considerable cost—the domesticated silkworm is no longer able to survive in the wild.
Humans today create domestic dependency in managed bees by our facilitation of the bees’ niche through our provision of an expanded nest cavity, the transporting of hives to better pasture or wintering areas, by our supplemental feeding of sugar and/or protein during dearth, our chemical control of varroa with miticides, and our medicating for other diseases. The question then is, how wimpy are those bees when forced to fend for themselves in an unforgiving natural environment–do escaped swarms from managed stock actually survive long enough in the wild to pass their genes into the wild populations of bees, and to what extent if they do?
Gene Flow Between Managed And Wild Stocks
There are a number of fully domesticated animals that, similar to the honey bee, are commonly expected to forage for themselves in the wild (and thus retain the potential to “go feral”). Some analogous examples would be sheep, goats, cattle, pigs, dogs, cats, chickens, ducks, or turkeys. In many areas, wild populations of these animals are able to interact and mate with domesticated animals of the same species.
So what happens when domesticated animals mate with wild stocks—do the genes of the domesticated animals introgress into the wild populations? This phenomenon, termed “genetic pollution,” is of great concern to wildlife biologists. So our question should be, in which direction do genes tend to flow—from domestic to wild, or the other way around?
As an example, let’s take a look at the wolf, which lives sympatrically with its domesticated form, the dog, in both Europe and North America (Fig. 2).

Figure 2. I ask you the rhetorical question, “How many Dalmatians would one need to introduce into the Canadian wolf population each year until you started to notice spotted, short-haired wolves?” Obviously, that’s not likely to happen, since despite the fact that Dalmations are about the same size, and fully able to hunt, wolves are far better adapted to the habitat and lifestyle (they exhibit greater fitness in the Canadian environment). Plus, as with other dog breeds, inbreeding has allowed deleterious alleles to accumulate–Dalmatians display a propensity towards deafness, allergies and urinary stones [23].
Despite the fact that domesticated dogs can mate with wolves (and wolves with coyotes, both resulting in fertile offspring), the races (and species) have long continued to remain genetically distinct. Even in the extreme case of the situation in Europe, where wolves (after being hunted nearly to extinction) have successfully recolonized dog-inhabited regions,
Genetic data showed that, despite occasional hybridization, wolf and dogs remain genetically distinct, suggesting that introgression in nature might be strongly counteracted by selection or by ethological factors [24].
The same researcher also studied the introgression of domesticated genes into wild populations of two other native European species—the European Wildcat (ancestral type of the domestic cat), and the introduced Chukar (related to the Red-Legged and Rock Partridges), and found that the degree of genetic introgression depends upon the amount of human influence in the environment, as well as the selective force of nature in the region [25].
He found that despite there being widespread occurrence of free-ranging domestic cats in Italy, the wildcat population remains genetically isolated. However, in some other countries there is sporadic hybridization; and in agriculturally-fragmented Scotland and Hungary, the wildcat population is deeply introgressed by domestic genetics. Similarly, captive-bred Chukars breed freely with wild partridges, but the survival of their offspring appears to be constrained by natural selection in alpine habitats.
Bottom line: it appears that wild-type populations of animals typically maintain genetic integrity despite an influx of domestic stocks, so long as nature applies strong selective pressure upon the population. However, those wishing to preserve the genetic purity of the races of Apis mellifera in their native ranges have every reason to be concerned about genetic pollution from introduced managed bees.
Back To Honey Bees
So, can established feral honey bee populations maintain their genetic integrity, despite an influx of swarms and drones from managed colonies? For an answer, we need only contemplate the legendarily successful invasion of Africanized bees (a feral hybrid of African and European races) in the New World. From a relatively small, but genetically diverse mixture of different original introductions, a well-adapted “superbee” was created, which quickly displaced all other honey bee races in the tropical and subtropical habitats of the Americas. Despite the continued introduction of domesticated European stocks into currently Africanized territory, the Africanized feral population shows no sign of being “diluted” by the domesticated bees.
The process of the genetic introgression of the Africanized bee into habitats to which they were better adapted than the preexisting (but poorly adapted) European races is nicely detailed by Pinto [26]. One should note that the Africanized bee has not displaced the European ferals in temperate areas of the Americas, to which the European races are better adapted (although I expect that there will eventually be some degree of genetic introgression).
Practical application: fitness in one environment does not imply fitness in all environments—a strong argument for propagating regionally-adapted stocks.
Along that line of thought, a study that years ago caught my eye (Daly 1991 [27]), was one of feral bees in California. The researchers wondered
…whether clinal [gradual] geographic variation [of the feral population] possibly could have developed during the past 138 years in the presence of a large, mobile beekeeping industry from which queens and drones must contribute annually to the gene pool of feral populations.
So they performed meticulous morphometric analysis [28] of 278 samples of feral bees and 49 samples of managed bees from all over the State, and compared them to those of known races of bees, as well as hive bees, from other states and countries . The results were fascinating. This was before Africanization, so all the samples appeared to derive from Central Mediterranean European races.
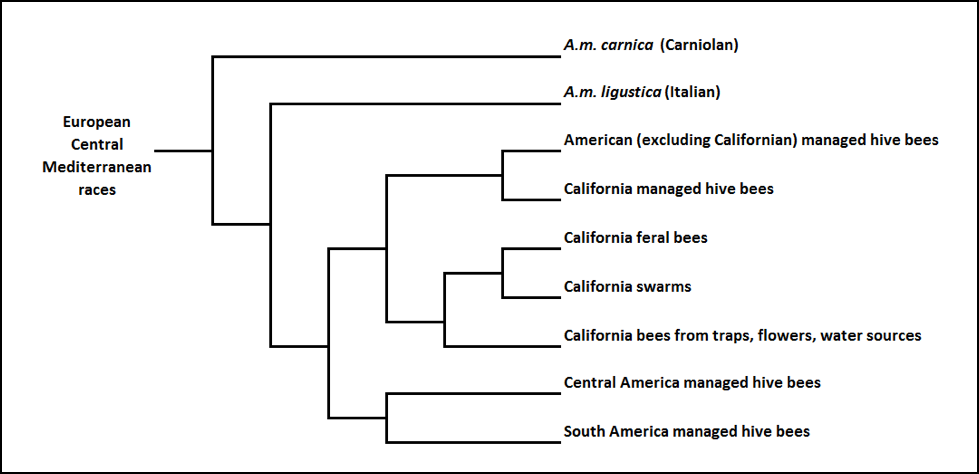
Their analysis segregated the sampled bees into different operational taxonomic units, as I’ve shown below (Fig. 3):
Figure 3. A phenogram [[i]] of the morphometric segregation of California feral and managed bees, relative to other bee populations. Note how all the samples had some similarity to pure Italians (not surprising in California’s Mediterranean climate), but were distinctly different. Then note the clear separation between the feral colonies and managed bees. Not shown are the African and Western Mediterranean reference samples, which segregated out separately. Phenogram reworked for clarity from Daly (1991).
[i] Phenogram: a diagram depicting taxonomic relationships among organisms based on overall similarity of many characteristics without regard to evolutionary history or assumed significance of specific characters: usually generated by computer. From Dictionary.com.
Of interest is that only 8% of the feral bees morphometrically resembled managed stocks. What I found most interesting (and hardly surprising) is that they also found (not shown) that the clinal variation of feral bees over the diverse California landscape appeared to be morphologically similar to native races of bees living in climatically similar European habitats.
Practical application: in the heart of U.S. bee keeping and breeding, feral populations of bees appeared to have evolved into established discrete regionally-adapted landraces [30] similar to those in Europe, despite what one would think to be an overwhelming influx of swarms and drones from the immense population of managed hives. Unfortunately, the ferals were decimated when varroa invaded California [31], but now appear to be slowly recovering [32].
A few years later, Schiff [33] genetically analyzed samples from 692 feral colonies from the southern U.S. and found that 37% had descended from the (long unpopular) Dark Bee (A.m. mellifera), and 2% had descended from an Egyptian race (A.m. lamarckii) imported prior to 1860. Schiff observed that:
The discovery of significant heterogeneity among U.S. feral honey bees implies that this population may represent a source of genetic variation for breeding programs [of great interest due to then concurrent invasion of varroa].
Practical application: the question then is, was the genetic diversity of the U.S. feral population lost when varroa wiped them out, or does the recovering feral population continue to retain its genetic diversity?
Drs. Steve Sheppard and Debbie Delaney have followed the genetic changes in the feral and domestic populations of honey bees since varroa. They are seeing what appears to be a recovery of genetically-distinct ferals throughout the country [34].
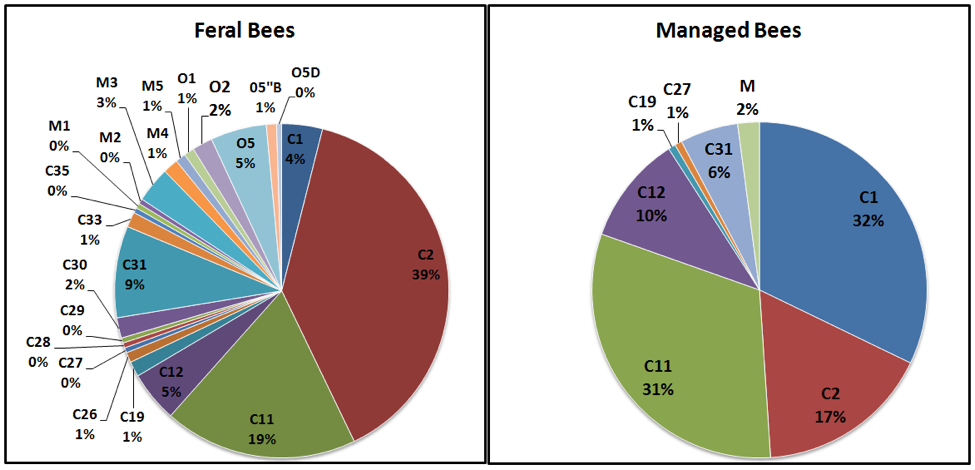
So what’s the bottom line? The most recent study by Roxane Magnus and Allen Szalanski [35] reviews previous studies, and appears to me to be the most up-to-date status report on the genetic diversity of U.S. honey bees. They analyzed samples of European worker bees collected (from 2005 – 2009) from 203 feral colonies and 44 swarms from 77 counties over 12 states, plus samples of 140 colonies from 14 queen breeders (their analysis of Africanized colonies was published separately [36]). They used a particularly sensitive method to analyze the mitochondrial DNA [37], which is inherited directly and solely through the maternal line. What they found was a diversity of markers (mitotypes) from 3 main lineages of European bees. Their results deserve close study (Fig. 4):
Figure 4. I put the data from Magnus [[i]] in graphical form, and labeled the name and percentage of the maternal lineages found in each group of sampled stocks. The various “C” lineages are from eastern European races, including Italian, Carniolan, and Caucasian stocks; these constituted 84% of the ferals and 98% of the managed bees. “M” is the Western European lineage, commonly known as the “German” or “Dark Bee”; found in 7% of the ferals, and 2% of managed. “O” is the Middle Eastern lineage (perhaps derived from long-ago importation of A.m. syriaca), found only in the ferals (about 10%).
[i] Magnus, R and AL Szalanski (2008) Genetic variation in honey bees from south central United States. Poster at 2008 ESA Open access.
Magnus, R & AL. Szalanski (2010) Genetic Evidence for Honey Bees (Apis mellifera L.) of Middle Eastern Lineage in the United States. Sociobiology 55(1B): 285-296.
Magnus, RM, et al (2011) Mitochondrial DNA diversity of honey bees, Apis mellifera L. (Hymenoptera: Apidae) form queen breeders in the United States. Journal of Apicultural Science 55(1): 37-46. http://www.jas.org.pl/jas_55_1_2011_4.pdf
Magnus, RM , AD Tripodi, AL. Szalanski (2014) Op. cit.
[i] Magnus, R and AL Szalanski (2008) Genetic variation in honey bees from south central United States. Poster at 2008 ESA Open access.
Magnus, R & AL. Szalanski (2010) Genetic Evidence for Honey Bees (Apis mellifera L.) of Middle Eastern Lineage in the United States. Sociobiology 55(1B): 285-296.
Magnus, RM, et al (2011) Mitochondrial DNA diversity of honey bees, Apis mellifera L. (Hymenoptera: Apidae) form queen breeders in the United States. Journal of Apicultural Science 55(1): 37-46. http://www.jas.org.pl/jas_55_1_2011_4.pdf
Magnus, RM , AD Tripodi, AL. Szalanski (2014) Op. cit.
Of note is the far greater genetic diversity of the feral bees (23 mitotypes) than in the managed bees (only 7 mitotypes total, with 82% of the diversity represented by only 3 mitotypes). Of interest is that the stocks of both western and southern queen producers tend to be predominated by only three C mitotypes [39], which likely flow into the feral populations (thus bolstering their proportions).
Practical food for thought: I find it of special interest that despite the fact that the C1 line (“yellow” Italian [40]) accounts for a third of the mitotypes in managed bees, that it does not appear to persist in the wild (only 4% of ferals carry it). Could it be that we tend to select for stock that lacks fitness under natural selective pressure?
The nomenclature and origins of the C2 group (which has a number of subgroups) is being sorted out [41], but appears to include Carniolan, Caucasian, and perhaps Cyprian stocks. Of interest is that this group was the most prevalent in the samples of ferals in this study. I can’t find where C11 comes from, but C12 may be Anatolian [42].
Also, what happened to the M line of “dark bees”? The U.S. feral population was originally founded solely from A.m. mellifera stock, but has since been largely supplanted by the C and O lines, which were introduced at later dates. So why is that? Was it solely due to the introgression pressure via escapees from managed stocks; or were the other lines simply better adapted to North American habitats? In answer, keep in mind that as of 1994, Schiff found that 37% of ferals to still retained M-line heritage, so the M line appeared to be able to hang in there despite massive introgression from managed stocks. Surprisingly then, by 2009 only 7% of Magnus’s feral samples were type M. What happened in the interim? Did varroa take the type M’s out? Or did Nosema ceranae [43]?
There is some evidence that some lineages are innately more resistant to certain parasites [44]. The question that haunts me is to what degree our recent elevated rate of colony mortality might be due to the limited gene pool of our managed stocks, which may simply be lacking critical genes for resistance to the onslaught of our recently-introduced parasites and the associated virus issues. Such a problem would likely be self correcting if it weren’t for the nearly universal reliance upon medications by our queen producers [45].
Perhaps of most practical application: a recent study by Tarpy [46] found that colonies that were less genetically diverse were 2.86 times more likely to die by the end of the study when compared to colonies that were more genetically diverse. This raises the serious question of whether our managed bees are a bit too inbred.
The above data certainly suggest that unbroken maternal lines of “survivor” bees appear to have persisted in the U.S. since their importations prior to 1922, despite having to recover from their devastation by varroa concurrent with beekeepers flooding the continent with roughly a million commercially-produced queens of domesticated stock each year.
Also of interest is that the authors make clear that:
It should be noted based on these results that mitochondrial DNA genetic variation does not reflect a specific phenotype; thus one mitotype may have multiple phenotypes.
This point deserves further explanation. Mitochondrial analysis is excellent for tracking genetic lineages (since it is only transmitted by the mother), but it’s not clear to what degree the random mutations in mitochondrial DNA affect the phenotypic appearance or performance of that line of bees [47]. Mitochondrial analysis does not take into account the genetics of the nuclear DNA, which codes for most of the proteins involved in an organism’s growth and behavior, and which is also transmitted by the drones, nor does it reflect epigenetic differences [48]. However, analysis of mitotypes is certainly useful as an indicator of overall genetic diversity and the persistence of mother lines in a population.
Practical application: What’s in a name? Pedigree may mean little to the phenotype which we observe in the honey bee. From a base pool of imported races, breeders can select for color or other characteristics atypical of the parental lines [49]. In the U.S., unless the parental line was recently imported (and maintained by instrumental insemination), any “breed” is likely mongrel stock selected for appearance and performance.
Bottom Line
In answer to my original questions:
- Yes, most managed breeds of honey bees can be considered to be domesticated (or at least semi-domesticated) animals, with their genetics largely controlled by human breeders rather than by nature.
- Domestication of an animal comes at a cost in fitness; i.e., a lessened ability to survive in the wild without human assistance (case in point, the C1 mitotype).
- Yes, there is truly a genetic difference between domesticated stocks and feral bees. Feral populations are not simply escaped swarms.
- Domesticated bees, by various mechanisms, may revert to a hybrid wild type that may be more genetically diverse and exhibit greater environmental fitness than the original parent races.
- Yes, U.S. feral bees maintain constantly-evolving, self-perpetuating populations (akin to “landraces” of other species, which tread the line between being wild, feral, and domesticated [50]). Strikingly, certain maternal lines appear to have persisted in the U.S. since at least the 1800’s, despite the introductions of chalkbrood, tracheal mite, Small Hive Beetle, Nosema ceranae, and who knows what else. The ferals are tough, and appear in many areas to be rebounding from the catastrophe of varroa.
- There is likely some degree of gene flow (mainly of C1 and C11) from the managed into the feral population, but appears to be little in return.
- Our American breeds of honey bees, be they called “Italians” or something else, are likely mongrels selected for particular characteristics and coloration, rather than genetic pedigree [51].
- I am in no way critical of our commercial queen producers, who deserve every penny for their excellent queens, which are typically bred for high productivity under best management practices.
- There is perhaps reason to be concerned about the paucity of genetic diversity in our managed bees. Every aspect of natural bee reproduction promotes genetic diversity; selective breeding from a limited gene pool goes against what has made the honey bee such a successful species.
- Our locally-adapted “survivor” feral populations may be an invaluable resource for bee breeders, offering the prospect of being our salvation from the varroa/virus complex.
- My strong feeling is that feral survivor stocks deserve far more of our attention than we have been giving them.
Acknowledgements
Thanks as always to Peter Borst for his generous assistance in my literature research, and to Dianne Behnke of Dadant for scanning old issues of ABJ. And thanks to those who donate to ScientificBeekeeping.com to allow me to continue my research and writing. And I thank my sons Eric and Ian for their patience as I barely squeaked out this article—written during an intensive April of California queenrearing and swarm management.
References And Footnotes
[1] My friend Dr. Ben Oldroyd would likely argue that I am stretching the term by referring our kept bees as being “domesticated.” In this article, I take the liberty of using the terms “managed” and “domesticated” interchangeably. Oldroyd, BP (2012) Domestication of honey bees was associated with expansion of genetic diversity. Molecular Ecology 21: 4409–4411. Open access.
[2] The term “phenotype” was not yet coined in Darwin’s time. The genotype-phenotype distinction was proposed by Wilhelm Johannsen in 1911 to make clear the difference between an organism’s heredity and what that heredity produces.
[3] Darwin, C (1875) The Variation of Animals and Plants under Domestication.
[4] Xia, Q, et al (2009) Complete resequencing of 40 genomes reveals domestication events and genes in silkworm (Bombyx). Science 326(5951): 433-436. Open access.
[5] From Wikipedia.
[6] This occurs when particular portions of the genotype are not expressed in the phenotype.
[7] The term “hybrid” has several different taxonomic meanings. It can refer to offspring resulting from the interbreeding of either (1) different species (or even genera), (2) different subspecies or races, or (3) different populations or domesticated breeds. All races of Apis mellifera readily hybridize.
[8] This observation should be obvious at face value, otherwise, the animal would in the worst case never make it past the embryo stage, or if it did, would be said to suffer from a genetic disease. See also Xia (2009) Op. cit.
[10] Chapman NC, J Lim, BP Oldroyd (2008) Population genetics of commercial and feral honey bees in western Australia. J Econ Entomol 101:272–277. Open access.
[11] Chapman NC, J Lim J & BP Oldroyd (2008) Population genetics of commercial and feral honey bees in Western Australia. Journal of Economic Entomology 101:272-277.
[12] Peacock, MM, et al (2009) Strong founder effects and low genetic diversity in introduced populations of Coqui frogs. Molecular Ecology 18: 3603–3615. Open access
[13] Sheppard WS (1989a) A history of the introduction of honey bee races into the United States; part I of a two-part series. Am Bee J 129:617–619.
Sheppard WS (1989b) A history of the introduction of honey bee races into the United States; part II of a two-part series. Am Bee J 129:664–667.
[14] Zeisset I and TJC Beebee (2003) Population genetics of a successful invader: the marsh frog Rana ridibunda. Britain. Molecular Ecology 12: 639–646.
[15] Because drones are produced only from unfertilized eggs, meaning that their genetics come only from their mothers.
[16] I’ll cover this topic more thoroughly in a later article.
[17] Weiser, EL, et al (2013) Simulating retention of rare alleles in small populations to assess management options for species with different life histories. Conservation Biology, 00(00): 1–10.
[18] Kolbe J, Glor R, Schettino L et al. (2004) Genetic variation increases during biological invasion by a Cuban lizard. Nature, 431, 177–181. Open access.
Suarez, AV and ND Tsutsui (2008) The evolutionary consequences of biological invasions. Molecular Ecology 17:351–360. Open access.
[19] Harpur, BA, et al (2012) Management increases genetic diversity of honey bees via admixture. Mol Ecol. 21(18):4414-21.
[20] Delaney, DA (2008) Genetic characterization of U.S. honey bee populations. PhD Dissertation, Washington State University. Open access.
[21] http://en.wikipedia.org/wiki/Domestication
[22] Hoy, MA (1976) Genetic Improvement of Insects: Fact or Fantasy. Environmental Entomology 5(5): 833-839.
[23] Images from Wikipedia and 1hdwallpapers.com
[24] Randi, E and V Lucchini (2002) Detecting rare introgression of domestic dog genes into wild wolf (Canis lupus) populations by Bayesian admixture analyses of microsatellite variation. Conservation Genetics 3(1): 29-43.
[25] Randi, E (2008) Detecting hybridization between wild species and their domesticated relatives. Molecular Ecology 17: 285-293. Open access.
[26] Pinto, MA, et al (2004) Temporal pattern of Africanization in a feral honeybee population from Texas inferred from mitochondrial DNA. Evolution 58:1047–1055. Open access.
[27] Daly, HV, et al (1991) Clinal geographic variation in feral honey bees in California, USA. Apidologie 22: 591-609. Open access.
[28] They compared 25 various body measurements.
[29] Phenogram: a diagram depicting taxonomic relationships among organisms based on overall similarity of many characteristics without regard to evolutionary history or assumed significance of specific characters: usually generated by computer. From Dictionary.com.
[30] Zeven, AC (1998). “Landraces: A review of definitions and classifications”. Euphytica 104 (2): 127–139.
[31] Kraus B and RE Page (1995) Effect of Varroa jacobsoni (Mesostigmata: Varroadae) on feral Apis mellifera (Hymenoptera: Apidae) in California. Environ Entomol. 24:80.
[32] Personal observations.
[33] Schiff, N. M., et al (1994) Genetic diversity of feral honey-bee (Hymenoptera, Apidae) populations in the southern United States. Ann. Entomol. Soc. Am. 87: 842-848.
[34] Dr. Deborah Delaney, pers. comm.
[35] Magnus, RM , AD Tripodi, AL. Szalanski (2014) Mitochondrial DNA Diversity of Honey Bees (Apis mellifera) from Unmanaged Colonies and Swarms in the United States. Biochemical Genetics [Epub ahead of print].
[36] Szalanski, AL and RM Magnus (2010) Mitochondrial DNA characterization of Africanized
honey bee (Apis mellifera L.) populations from the USA. Journal of Apicultural Research and Bee World 49(2): 177-185
[37] For example, taxonomical classification by mitotype does not always jibe with that of morphometrics:
Smith, DR (1991) African bees in the Americas: Insights from biogeography and genetics. Trends Ecol Evol. 6(1):17-21. Open access.
[38] Magnus, R and AL Szalanski (2008) Genetic variation in honey bees from south central United States. Poster at 2008 ESA Open access.
Magnus, R & AL. Szalanski (2010) Genetic Evidence for Honey Bees (Apis mellifera L.) of Middle Eastern Lineage in the United States. Sociobiology 55(1B): 285-296.
Magnus, RM, et al (2011) Mitochondrial DNA diversity of honey bees, Apis mellifera L. (Hymenoptera: Apidae) form queen breeders in the United States. Journal of Apicultural Science 55(1): 37-46. http://www.jas.org.pl/jas_55_1_2011_4.pdf
Magnus, RM , AD Tripodi, AL. Szalanski (2014) Op. cit.
[39] Delaney, DA (2008) Op. cit.
[40] Oxley, PR and BP Oldroyd (2009) Mitochondrial sequencing reveals five separate origins of ‘black’ Apis mellifera (Hymenoptera: Apidae) in eastern Australian commercial colonies. Journal of Economic Entomology 102(2):480-484.
[41] Nedić, N, et al (2009) Molecular characterization of the honeybee Apis mellifera carnica In Serbia. Arch. Biol. Sci., Belgrade, 61 (4), 587-598.
[42] Solorzano, CD, et al (2009) Phylogeography and Population Genetics of Honey Bees (Apis mellifera) From Turkey Based on COI-COII Sequence Data. Sociobiology Vol. 53, No. 1: 237-246. Open access.
[43] Whitaker, J, et al (2011) Molecular detection of Nosema ceranae and N. apis from Turkish honey bees. Apidologie 42:174–180 (read the discussion). Open access.
[44] Jensen, AB, et al (2009) Differential susceptibility across honey bee colonies in larval chalkbrood resistance. Apidologie 40: 524–534. Open access.
[45] Myself included–I still find it necessary to treat for varroa. However, it is difficult to select for naturally-resistant stock if one routinely applies prophylactic antibiotic treatments.
[46] Tarpy, DR, D vanEngelsdorp, JS Pettis (2013) Genetic diversity affects colony survivorship in commercial honey bee colonies. Naturwissenschaften 100(8): 723-728.
[47] The differences and applicability of the classification methods are well explained by:
Darger, K (2013) Determining low levels of Africanization in unmanaged honey bee colonies using three diagnostic techniques. M.S. Thesis, Univ of Delaware. Open access.
[48] Mitochondria are cellular organelles that convert sugar to energy, as well as some other critical functions. Mitochondria have their own DNA independent of the cell’s nuclear DNA, which codes for all the other proteins for body growth and functioning. The assumption is that the egg destroys any paternal mitochondria from the sperm, but there may be exceptions, although the odds are that mitotyping would be accurate at tracking maternal geneology.
[49] Oxley, PR and BP Oldroyd (2009) Op. cit.
[50] http://en.wikipedia.org/wiki/Landrace
[51] Darger (2013) Op. cit.